| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,230,694 – 5,230,788 |
| Length | 94 |
| Max. P | 0.999978 |

| Location | 5,230,694 – 5,230,788 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 58.75 |
| Shannon entropy | 0.72704 |
| G+C content | 0.49844 |
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -14.67 |
| Energy contribution | -14.20 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.53 |
| Mean z-score | -4.42 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.57 |
| SVM RNA-class probability | 0.999978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
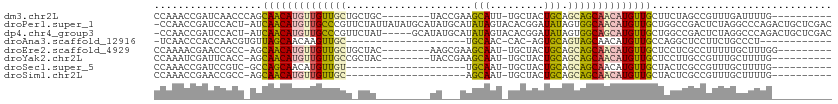
>dm3.chr2L 5230694 94 + 23011544 CCAAACCGAUCAACCCAGCAACAUGUUGUUGCUGCUGC--------UACCGAAGCAUU-UGCUACUGCAGCAGCAACAUGUUGCUUCUAGCCGUUUGAUUUUG---------- .(((((.(........((((((((((((((((((((((--------(.....))))..-.......))))))))))))))))))).....).)))))......---------- ( -37.82, z-score = -5.87, R) >droPer1.super_1 8469991 111 + 10282868 -CCAACCGAUCCACU-AUCAACAUGUUGCCCGUUCUAUUAUAUGCAUAUGCAUAUAGUACACGGAUAUAGUGGCAACAUGUUGCUGGCCGACUCUAGGCCCAGACUGCUCGAC -.....(((......-..(((((((((((((((..((((((((((....)))))))))).)))........))))))))))))((((((.......)).)))).....))).. ( -39.00, z-score = -4.16, R) >dp4.chr4_group3 7017699 106 + 11692001 -CCAACCGAUCCACU-AUCAACAUGUUGCCCGUUCUAU-----GCAUAUGCAUAUAGUACACGGAUAUAGUGGCAGCAUGUUGCUGGCCGACUCUAGGCCCAGACUGCUCGAC -.....(((......-..((((((((((((.((..(((-----((....)))))....))((.......))))))))))))))((((((.......)).)))).....))).. ( -32.50, z-score = -1.63, R) >droAna3.scaffold_12916 6247564 78 - 16180835 -UCAACCCACCAACGUGUUAGCAACAAGUUGC--------------------UGCAAC-CAC-AGUGCAGUAGCAACAUGUUGCCAGGCUCCUUCUGCCCU------------ -......(((....)))...((((((.(((((--------------------(((.((-...-.))...)))))))).))))))..(((.......)))..------------ ( -24.40, z-score = -2.46, R) >droEre2.scaffold_4929 5309548 94 + 26641161 CCAAAACGAACCGCC-AGCAACAUGUUGUUGCUGCUAC--------AAGCGAAGCAAU-UGCUACUGCAGCAGCAACAUGUUGCUCCUCGCCUUUUUGCUUUGG--------- (((((.((((..((.-(((((((((((((((((((...--------.(((((.....)-))))...)))))))))))))))))))....))...)))).)))))--------- ( -43.70, z-score = -6.45, R) >droYak2.chr2L 8356678 93 - 22324452 CCAAAUCGAUUCACC-AGCAACAUGUUGUUGCCGCUAC--------UACCGAAGCAAU-UGCUACUGCAGCAGCAACAUGUUGCUCCUUGCCGUUUGCUUUUG---------- .(((((.(.......-((((((((((((((((.(((..--------......)))...-(((....))))))))))))))))))).....).)))))......---------- ( -32.60, z-score = -4.53, R) >droSec1.super_5 3311446 81 + 5866729 CCAAACCGAUCCGUC-GCCAGCAACAUGUUGU--------------------UGCAAU-UGCUACUGCAGCAGCAACAUGUUGCUACUCGCCGUUUGCUUUUG---------- .((((((((....))-)..(((((((((((((--------------------(((...-(((....))))))))))))))))))).......)))))......---------- ( -32.30, z-score = -4.93, R) >droSim1.chr2L 5029190 81 + 22036055 CCAAACCGAACCGCC-AGCAACAUGUUGUUGC--------------------AGCAAU-UGCUACUGCAGCAGCAACAUGUUGCUACUCGCCGUUUGCUUUUG---------- .((((.(((((.((.-((((((((((((((((--------------------.(((..-......))).))))))))))))))))....)).)))))..))))---------- ( -35.10, z-score = -5.33, R) >consensus CCAAACCGAUCCACC_AGCAACAUGUUGUUGCU_CUA_________UA___AAGCAAU_UGCUACUGCAGCAGCAACAUGUUGCUACUCGCCGUUUGCCUUUG__________ ..................((((((((((((((.....................(((.........))).)))))))))))))).............................. (-14.67 = -14.20 + -0.47)
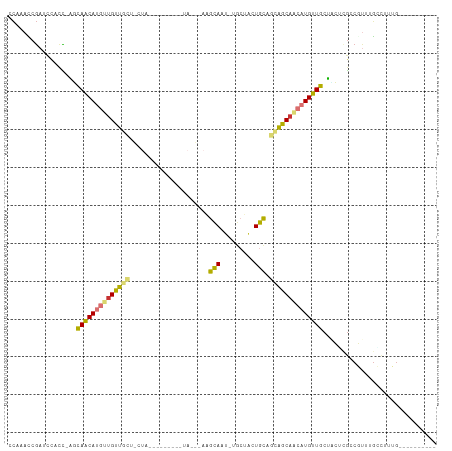
| Location | 5,230,694 – 5,230,788 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 58.75 |
| Shannon entropy | 0.72704 |
| G+C content | 0.49844 |
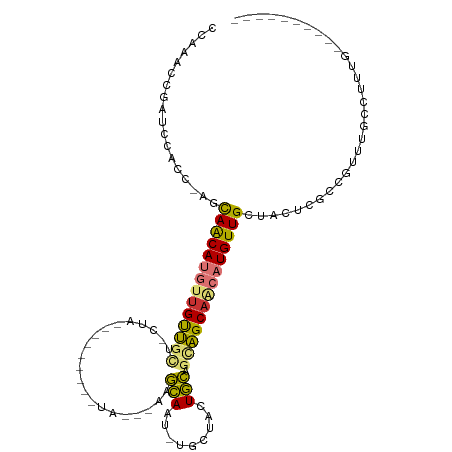
| Mean single sequence MFE | -35.86 |
| Consensus MFE | -5.60 |
| Energy contribution | -4.92 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.58 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.16 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5230694 94 - 23011544 ----------CAAAAUCAAACGGCUAGAAGCAACAUGUUGCUGCUGCAGUAGCA-AAUGCUUCGGUA--------GCAGCAGCAACAACAUGUUGCUGGGUUGAUCGGUUUGG ----------..(((((...(((((...(((((((((((((((((((.(.(((.-...))).)....--------)))))))))...)))))))))).)))))...))))).. ( -40.90, z-score = -4.83, R) >droPer1.super_1 8469991 111 - 10282868 GUCGAGCAGUCUGGGCCUAGAGUCGGCCAGCAACAUGUUGCCACUAUAUCCGUGUACUAUAUGCAUAUGCAUAUAAUAGAACGGGCAACAUGUUGAU-AGUGGAUCGGUUGG- ..(((.(.((((.((((.......))))..((((((((((((........(((.((.(((((((....))))))).))..)))))))))))))))..-...)))).).))).- ( -40.30, z-score = -2.51, R) >dp4.chr4_group3 7017699 106 - 11692001 GUCGAGCAGUCUGGGCCUAGAGUCGGCCAGCAACAUGCUGCCACUAUAUCCGUGUACUAUAUGCAUAUGC-----AUAGAACGGGCAACAUGUUGAU-AGUGGAUCGGUUGG- .(((((((((...((((.......))))....)).)))..(((((((.(((((......(((((....))-----)))..)))))(((....)))))-))))).))))....- ( -33.10, z-score = -0.18, R) >droAna3.scaffold_12916 6247564 78 + 16180835 ------------AGGGCAGAAGGAGCCUGGCAACAUGUUGCUACUGCACU-GUG-GUUGCA--------------------GCAACUUGUUGCUAACACGUUGGUGGGUUGA- ------------.((((.......))))(((((((((((((.((..(...-)..-)).)))--------------------)))...)))))))..(((....)))......- ( -26.50, z-score = -0.33, R) >droEre2.scaffold_4929 5309548 94 - 26641161 ---------CCAAAGCAAAAAGGCGAGGAGCAACAUGUUGCUGCUGCAGUAGCA-AUUGCUUCGCUU--------GUAGCAGCAACAACAUGUUGCU-GGCGGUUCGUUUUGG ---------(((((((.((...((.((.((((...((((((((((((((((((.-...)))..)).)--------))))))))))))...)))).))-.))..)).))))))) ( -42.20, z-score = -3.84, R) >droYak2.chr2L 8356678 93 + 22324452 ----------CAAAAGCAAACGGCAAGGAGCAACAUGUUGCUGCUGCAGUAGCA-AUUGCUUCGGUA--------GUAGCGGCAACAACAUGUUGCU-GGUGAAUCGAUUUGG ----------......((((((.((...(((((((((((((((((((...(((.-...)))...)))--------)))))(....))))))))))))-..))...)).)))). ( -37.50, z-score = -3.85, R) >droSec1.super_5 3311446 81 - 5866729 ----------CAAAAGCAAACGGCGAGUAGCAACAUGUUGCUGCUGCAGUAGCA-AUUGCA--------------------ACAACAUGUUGCUGGC-GACGGAUCGGUUUGG ----------((((......((.((..(((((((((((((.((((((....)))-...)))--------------------.))))))))))))).)-).))......)))). ( -31.10, z-score = -2.76, R) >droSim1.chr2L 5029190 81 - 22036055 ----------CAAAAGCAAACGGCGAGUAGCAACAUGUUGCUGCUGCAGUAGCA-AUUGCU--------------------GCAACAACAUGUUGCU-GGCGGUUCGGUUUGG ----------((((..(.(((.((...(((((((((((((.(((.(((((....-))))).--------------------))).))))))))))))-))).))).).)))). ( -35.30, z-score = -3.39, R) >consensus __________CAAAAGCAAAAGGCGAGUAGCAACAUGUUGCUGCUGCAGUAGCA_AUUGCAU___UA_________UAG_AGCAACAACAUGUUGCU_GGUGGAUCGGUUUGG .....................(((((...((((....))))(((.......)))......................................)))))................ ( -5.60 = -4.92 + -0.67)

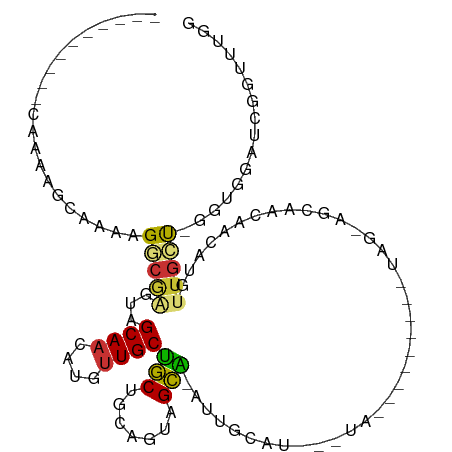
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:22 2011