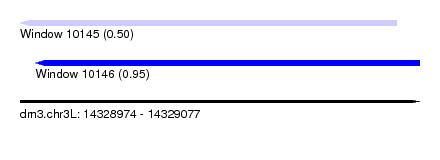
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,328,974 – 14,329,077 |
| Length | 103 |
| Max. P | 0.947878 |

| Location | 14,328,974 – 14,329,071 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 82.26 |
| Shannon entropy | 0.27494 |
| G+C content | 0.44190 |
| Mean single sequence MFE | -19.43 |
| Consensus MFE | -14.25 |
| Energy contribution | -13.63 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3L 14328974 97 - 24543557 AUCCCCUUUUAGAAUCUCCGCCUCUGGACUUAGGCCUAGACAAGUUCUACUUGUUUUGACUGCUCUUGUCUUCCUCGCCUCCCUAUAUUUUUUUUUU .........(((....((((....))))...((((..(((((((.....))))))).(((.......)))......))))..)))............ ( -14.60, z-score = -1.04, R) >droSim1.chr3L 13675168 97 - 22553184 AUCCCCUUUUAGAAUCUAAGCCGCUGAACUUAGGCCUAGACAAGUUCUACUUGUUUGGACUGCUCUUGUCUUCCUCGCCUCCCUAUGUUUUUUUUUU .........................((((.((((...(((((((.....)))))))(((..((....))..))).......)))).))))....... ( -15.40, z-score = -0.33, R) >droYak2.chr3L 14418841 90 - 24197627 UCCCCCUUUUAGAAGCUCCACCUCUGGAUUUAGGUCUAGACAAGUUCUACUUGUUUGGACUGCUUUUGUCUUCUUCACCUCCCUCUGGUU------- .........(((((((((((....))))....((((((((((((.....)))))))))))))))))))........(((.......))).------- ( -26.10, z-score = -3.03, R) >droEre2.scaffold_4784 14311257 90 - 25762168 UCCCCUUUUCGGAAUCUCCGCCUCUGGAUUUAGGCCUAGACAAGUUCUACUUGUUUGGAUAGCUUUUGUCUUCUUCGCCUCCCUCUGAUU------- ........(((((......((....((((..(((((((((((((.....)))))))))...))))..)))).....)).....)))))..------- ( -21.60, z-score = -1.58, R) >consensus ACCCCCUUUUAGAAUCUCCGCCUCUGGACUUAGGCCUAGACAAGUUCUACUUGUUUGGACUGCUCUUGUCUUCCUCGCCUCCCUAUGUUU_______ .......((((((.........))))))...((((.((((((((.....))))))))(((.......)))......))))................. (-14.25 = -13.63 + -0.62)

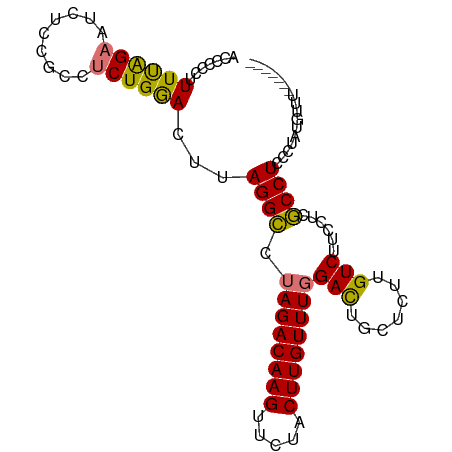
| Location | 14,328,978 – 14,329,077 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 66.83 |
| Shannon entropy | 0.62619 |
| G+C content | 0.45214 |
| Mean single sequence MFE | -21.19 |
| Consensus MFE | -11.77 |
| Energy contribution | -11.67 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
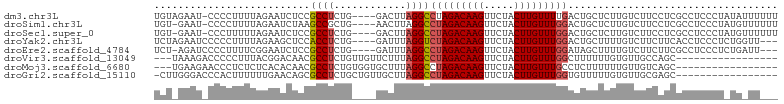
>dm3.chr3L 14328978 99 - 24543557 UGUAGAAU-CCCCUUUUAGAAUCUCCGCCUCUG----GACUUAGGCCUAGACAAGUUCUACUUGUUUUGACUGCUCUUGUCUUCCUCGCCUCCCUAUAUUUUUU (((((...-..............((((....))----))...((((..(((((((.....))))))).(((.......)))......))))..)))))...... ( -16.80, z-score = -0.75, R) >droSim1.chr3L 13675172 98 - 22553184 UGU-GAAU-CCCCUUUUAGAAUCUAAGCCGCUG----AACUUAGGCCUAGACAAGUUCUACUUGUUUGGACUGCUCUUGUCUUCCUCGCCUCCCUAUGUUUUUU .((-((..-............((((((......----..)))))).(((((((((.....)))))))))................))))............... ( -16.60, z-score = -0.05, R) >droSec1.super_0 6465904 98 - 21120651 UGU-GAAU-CCCUUUUUAGAAUCUCCGCCUCUG----GACUUAGGCCUAGACAAGUUCUACUUGUUUGGACUGCUCUUGUCUUCCUCGCCUCCCUAUGUUUUUU .((-((..-.......((((.........))))----(((...((((((((((((.....)))))))))...)))...)))....))))............... ( -17.70, z-score = -0.38, R) >droYak2.chr3L 14418841 97 - 24197627 UCUAGAAUCCCCCUUUUAGAAGCUCCACCUCUG----GAUUUAGGUCUAGACAAGUUCUACUUGUUUGGACUGCUUUUGUCUUCUUCACCUCCCUCUGGUU--- ................(((((((((((....))----))....((((((((((((.....)))))))))))))))))))........(((.......))).--- ( -26.10, z-score = -2.17, R) >droEre2.scaffold_4784 14311257 96 - 25762168 UCU-AGAUCCCCUUUUCGGAAUCUCCGCCUCUG----GAUUUAGGCCUAGACAAGUUCUACUUGUUUGGAUAGCUUUUGUCUUCUUCGCCUCCCUCUGAUU--- ..(-((((((......(((.....))).....)----))))))((((((((((((.....)))))))))...((....)).......)))...........--- ( -22.90, z-score = -1.20, R) >droVir3.scaffold_13049 22011032 84 - 25233164 ---UAAAGACCCCCUUUACGGACAACGCCUCUGUUGUUCUUUAGGCCUAGACAAGUUCUACUUGUUUGGCUUUUUUGUGUUGCCAGC----------------- ---(((((.....))))).((.((((((..(((........)))(((.(((((((.....))))))))))......))))))))...----------------- ( -23.00, z-score = -2.16, R) >droMoj3.scaffold_6680 16576432 84 - 24764193 ---UGAAGAACCCUCUCUCACACAACGCCUCUGUGGUGCUUUAGGCCUAGACAAGUUCUACUUGUUUGCCUCUUUUUUGUUGUCAGC----------------- ---((((((....))).))).((((((((.....))).....((((..(((((((.....))))))))))).......)))))....----------------- ( -18.90, z-score = -0.61, R) >droGri2.scaffold_15110 3017753 86 + 24565398 -CUUGGGACCCACUUUUUUGAACAGCGCCUCUGCUGUUGCUUAGGCCUAGACAAGUUCUACUUGUUUGGUGUUUUUGUGUUGCGAGC----------------- -((((.(((...........(((((((....)))))))((..(((((((((((((.....))))))))).))))..))))).)))).----------------- ( -27.50, z-score = -2.08, R) >consensus UCU_GAAU_CCCCUUUUAGAAUCUCCGCCUCUG____GACUUAGGCCUAGACAAGUUCUACUUGUUUGGACUGCUUUUGUCUUCAUCGCCUCCCU_UG_UU___ ..........................((((..(......)..))))(((((((((.....)))))))))................................... (-11.77 = -11.67 + -0.09)
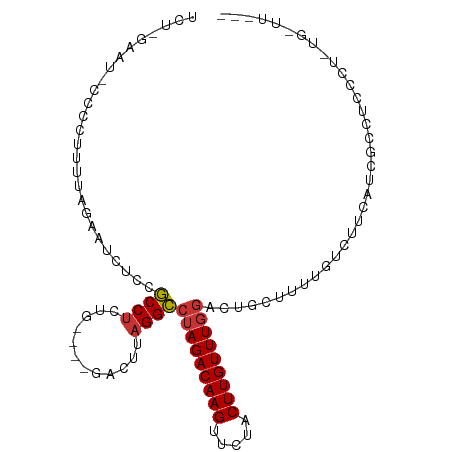
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:36 2011