
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,256,268 – 14,256,328 |
| Length | 60 |
| Max. P | 0.912100 |

| Location | 14,256,268 – 14,256,328 |
|---|---|
| Length | 60 |
| Sequences | 8 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 68.10 |
| Shannon entropy | 0.58649 |
| G+C content | 0.56578 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -9.44 |
| Energy contribution | -10.33 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
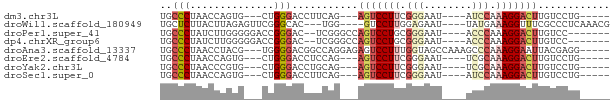
>dm3.chr3L 14256268 60 + 24543557 UGCCCUAACCAGUG---CUGGGACCUUCAG---AGUCCUUCGGGAAU----AUCCAAAGGACUUGUCCUG----- .((.((....)).)---).(((((.....(---(((((((.((....----..)).))))))))))))).----- ( -19.10, z-score = -1.41, R) >droWil1.scaffold_180949 3551372 64 + 6375548 UGCUCUUACUUAGAGUUCGGGCAC---UGG----GUCCUUGGAGAAU----UAUGAAAGGUUUCGCCCUCAAACG .(((((.....)))))..((((..---.((----(.((((.......----.....)))).)))))))....... ( -14.30, z-score = 0.29, R) >droPer1.super_41 304598 62 + 728894 UGCCCUAUCUUGGGGGACCGGGAC--UCGGGCCAGUCCUGCGGGAAU----ACCCAAAGGACUUGUCC------- .((((..((((((....)))))).--..)))).((((((..(((...----.)))..)))))).....------- ( -30.10, z-score = -2.36, R) >dp4.chrXR_group6 6943750 62 + 13314419 UGCCCUAUCUUGGGGGACCGGGAC--UCGGGCCAGUCCUGCGGGAAU----ACCCAAAGGACUUGUCC------- .((((..((((((....)))))).--..)))).((((((..(((...----.)))..)))))).....------- ( -30.10, z-score = -2.36, R) >droAna3.scaffold_13337 18497628 67 - 23293914 UGCCCUAACCUACG---UGGGGACGGCCAGGAGAGUCCUUUGGUAGCCAAAGCCCAAAGGAAUUACGAGG----- ........(((.((---(((((....)).......((((((((..((....)))))))))).))))))))----- ( -18.80, z-score = 0.20, R) >droEre2.scaffold_4784 14247667 60 + 25762168 UGCCCUAACCAGUG---CUGGGACCUCCAG---AGUCCUUCGGGAAU----UCGCAAAGGACUUGUCCUG----- .((.((....)).)---).(((((.....(---(((((((.(.(...----.).).))))))))))))).----- ( -18.20, z-score = -0.74, R) >droYak2.chr3L 14354593 60 + 24197627 UGCCCUAACCCGUG---CUGGGACCUGCAG---AGUCCUUCGGGAAU----UCGCAAAGGACUUGUCCUG----- (((.....((((..---..((((((....)---.))))).))))...----..))).((((....)))).----- ( -19.50, z-score = -1.25, R) >droSec1.super_0 6403479 60 + 21120651 UGCCCUAACCAGUG---CUGGGACCUUCAG---AGUCCUUCGGGAAU----AUCCAAAGGACUUGUCCUG----- .((.((....)).)---).(((((.....(---(((((((.((....----..)).))))))))))))).----- ( -19.10, z-score = -1.41, R) >consensus UGCCCUAACCUGUG___CUGGGACCUUCAG___AGUCCUUCGGGAAU____ACCCAAAGGACUUGUCCUG_____ ..(((..............)))...........(((((((.(((........))).)))))))............ ( -9.44 = -10.33 + 0.89)
| Location | 14,256,268 – 14,256,328 |
|---|---|
| Length | 60 |
| Sequences | 8 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 68.10 |
| Shannon entropy | 0.58649 |
| G+C content | 0.56578 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -12.71 |
| Energy contribution | -13.90 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
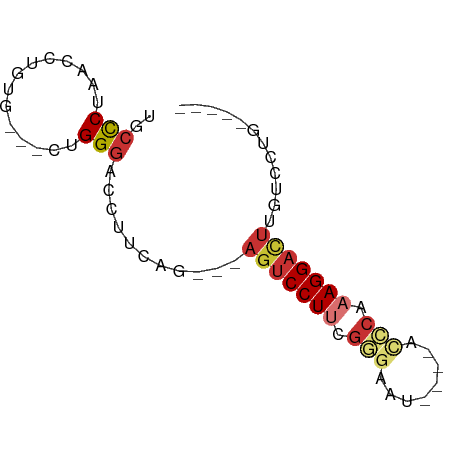
>dm3.chr3L 14256268 60 - 24543557 -----CAGGACAAGUCCUUUGGAU----AUUCCCGAAGGACU---CUGAAGGUCCCAG---CACUGGUUAGGGCA -----(((....((((((((((..----....))))))))))---)))...((((.((---(....))).)))). ( -23.10, z-score = -2.02, R) >droWil1.scaffold_180949 3551372 64 - 6375548 CGUUUGAGGGCGAAACCUUUCAUA----AUUCUCCAAGGAC----CCA---GUGCCCGAACUCUAAGUAAGAGCA .......(((((...((((.....----.......))))..----...---.)))))...((((.....)))).. ( -11.90, z-score = 0.23, R) >droPer1.super_41 304598 62 - 728894 -------GGACAAGUCCUUUGGGU----AUUCCCGCAGGACUGGCCCGA--GUCCCGGUCCCCCAAGAUAGGGCA -------.....((((((.((((.----...)))).)))))).((((..--(((..((....))..))).)))). ( -25.70, z-score = -1.44, R) >dp4.chrXR_group6 6943750 62 - 13314419 -------GGACAAGUCCUUUGGGU----AUUCCCGCAGGACUGGCCCGA--GUCCCGGUCCCCCAAGAUAGGGCA -------.....((((((.((((.----...)))).)))))).((((..--(((..((....))..))).)))). ( -25.70, z-score = -1.44, R) >droAna3.scaffold_13337 18497628 67 + 23293914 -----CCUCGUAAUUCCUUUGGGCUUUGGCUACCAAAGGACUCUCCUGGCCGUCCCCA---CGUAGGUUAGGGCA -----....((...((((((((((....))..))))))))....(((((((.......---....))))))))). ( -22.20, z-score = -0.69, R) >droEre2.scaffold_4784 14247667 60 - 25762168 -----CAGGACAAGUCCUUUGCGA----AUUCCCGAAGGACU---CUGGAGGUCCCAG---CACUGGUUAGGGCA -----(((....(((((((((.(.----...).)))))))))---)))...((((.((---(....))).)))). ( -19.90, z-score = -0.56, R) >droYak2.chr3L 14354593 60 - 24197627 -----CAGGACAAGUCCUUUGCGA----AUUCCCGAAGGACU---CUGCAGGUCCCAG---CACGGGUUAGGGCA -----(((....(((((((((.(.----...).)))))))))---)))...((((.((---(....))).)))). ( -19.00, z-score = -0.49, R) >droSec1.super_0 6403479 60 - 21120651 -----CAGGACAAGUCCUUUGGAU----AUUCCCGAAGGACU---CUGAAGGUCCCAG---CACUGGUUAGGGCA -----(((....((((((((((..----....))))))))))---)))...((((.((---(....))).)))). ( -23.10, z-score = -2.02, R) >consensus _____CAGGACAAGUCCUUUGGGU____AUUCCCGAAGGACU___CUGAAGGUCCCAG___CACAGGUUAGGGCA .......((...((((((((((..........))))))))))...))....((((...............)))). (-12.71 = -13.90 + 1.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:26 2011