| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,226,993 – 14,227,092 |
| Length | 99 |
| Max. P | 0.792126 |

| Location | 14,226,993 – 14,227,089 |
|---|---|
| Length | 96 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.85 |
| Shannon entropy | 0.72812 |
| G+C content | 0.45255 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -7.33 |
| Energy contribution | -7.30 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 2.17 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792126 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
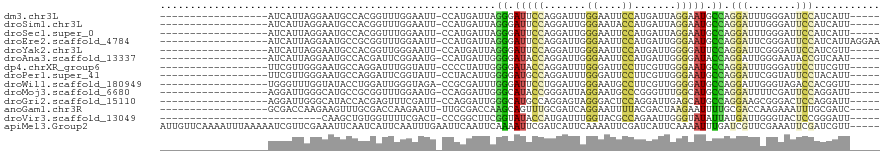
>dm3.chr3L 14226993 96 - 24543557 ------------------AUCAUUAGGAAUGCCACGGUUUGGAAUU-CCAUGAUUAGGGAUUCCAGGAUUUGGAAUUCCAUGAUUAGGAAUGCCAGGAUUUGGGAUUCCAUCAUU----- ------------------.......(((((.(((...(((((.(((-((.((((((((((((((((...)))))))))).))))))))))).)))))...))).)))))......----- ( -39.40, z-score = -4.14, R) >droSim1.chr3L 13582239 96 - 22553184 ------------------AUCAUUAGGAAUGCCACGGUUUGGAAUU-CCAUGAUUAGGGAUUCCAGGAUUGGGAAUACCAUGAUUAGGAAUGCCAGGAUUUGGGAUUCCAUCAUU----- ------------------.......(((((.(((...(((((.(((-((.((((((.(((((((.......))))).)).))))))))))).)))))...))).)))))......----- ( -34.80, z-score = -2.88, R) >droSec1.super_0 6372845 96 - 21120651 ------------------AUCAUUAGGAAUGCCACGGUUUGGAAUU-CCAUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUGAUUAGGAAUGCCAGGAUUUGGGAUUCCAUCAUU----- ------------------.......(((((.(((...(((((.(((-((.((((((((((((((.......)))))))).))))))))))).)))))...))).)))))......----- ( -38.40, z-score = -3.64, R) >droEre2.scaffold_4784 14219166 101 - 25762168 ------------------AUCAUUAGGAAUGCCGCGGUUUGGAAUU-CCAUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUGAUUGGGAAUGCCAGGAUUCGGGAUUCCAUCAUUAGGAA ------------------.......(((((.(((...(((((.(((-((.((((((((((((((.......)))))))).))))))))))).)))))...))).)))))........... ( -38.10, z-score = -2.62, R) >droYak2.chr3L 14324561 96 - 24197627 ------------------AUCAUUAGGAAUGCCACGGUUGGGAAUU-CCAUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUGAUUGGGGAUUCCAGGAUUCGGGAUUCCAUCGUU----- ------------------.......(((((.((.......((((((-((.......)))))))).(((((.((((((((.......))))))))..))))))).)))))......----- ( -37.20, z-score = -2.33, R) >droAna3.scaffold_13337 18466010 96 + 23293914 ------------------AUCAUUAGGAAUGCCACGAUUCGGAAUG-CCAUGAUUGGGGAUACCAGGAUUGGGAAUUCCAUGAUUGGGGAUACCAGGAUUGGGAAUACCGUCAAU----- ------------------.......((.((.((.(((((((((((.-(((...((((.....))))...)))..))))).....(((.....))))))))))).)).))......----- ( -28.20, z-score = -0.69, R) >dp4.chrXR_group6 6916498 96 - 13314419 ------------------UUCGUUGGGAAUGCCAGGAUUUGGUAUU-CCCCUAUUGGGGAUACCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUUGGGAUUCCUUCGUU----- ------------------..((..((((((.(((((.((((((((.-((((....)))))))))))).(((((.((((((.....)))))).))))).))))).)))))).))..----- ( -44.10, z-score = -3.81, R) >droPer1.super_41 274448 96 - 728894 ------------------UUCGUUGGGAAUGCCAGGAUUCGGUAUU-CCUACAUUGGGGAUGCCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUCGGUAUUCCUACAUU----- ------------------...((.(((((((((.((((((((((((-((((....((((((.((((...)))).))))))....)))))))))).)))))))))))))))))...----- ( -46.60, z-score = -5.49, R) >droWil1.scaffold_180949 3514038 96 - 6375548 ------------------UGGGUUUGGUAUACCUGGAUUGGGUAGA-CCGCGAUUUGGGAUUCCUGGAUUGGGAAUGCCUUCGUUGGGGAUGCCAGGAUUGGGUAGACCACGGUU----- ------------------......((((.((((..(.(..((((..-((((((...((.((((((.....)))))).)).))))..))..))))..).)..)))).)))).....----- ( -37.10, z-score = -2.62, R) >droMoj3.scaffold_6680 8059378 96 + 24764193 ------------------AGGAUUGGGCAUGCCGCGGUUUGGAAUG-CCAGGAUUGGGCAUACCGGGAUUAGGAAUGCCCGGGUUUGGCAUGCCAGGAUUUUCGAUUCCAGGAUU----- ------------------.......((((((((..(((.......)-))....((((((((.((.......)).))))))))....)))))))).(((........)))......----- ( -37.50, z-score = -1.97, R) >droGri2.scaffold_15110 21437788 96 - 24565398 ------------------AGGAUUGGGCAUACCACGAGUUUCGAUU-CCAGGAUUGGGCAUGCCAGGAGUAGGGACUCCAGGAUUGAGCAUGCCAGGAAGCGGGACUCCAGGAUU----- ------------------.....(((.....))).((((..((.((-((.......((((((((((((((....))))).....)).))))))).)))).))..)))).......----- ( -34.90, z-score = -2.24, R) >anoGam1.chr3R 30836173 96 + 53272125 ------------------GCGACCAAGAAGUUUGCGACCAAGAAUU-UUGCGACCAAGCAGUUUGCGAUCAGGAAUUUUACGACUAAGAAUUUUGCGACCAAGAAAUUUGCGAUC----- ------------------((((.(..(((((((........)))))-))((......)).).))))((((.((((((((.......))))))))((((.........))))))))----- ( -14.90, z-score = 0.96, R) >droVir3.scaffold_13049 3121581 87 + 25233164 ---------------------------CAAGCUGUGGUUUUCGACU-CCCGGCUUCGGUAUACCAUGAUUUGGUACGCCAGAAUUGGGUAUAUUAUGAUUGGGUACUCCGGGAUU----- ---------------------------.((((((.((.........-))))))))(((..((((.((((((((((..((.......))..))))).)))))))))..))).....----- ( -23.50, z-score = -0.15, R) >apiMel3.Group2 14092252 115 + 14465785 AUUGUUCAAAAUUUAAAAAUCGUUCGAAAUUCAAUCAUUCAAUUUGAAUUCAAUUCAAAAUUCGAUCAUUCAAAAUUCGAUCAUUCAAAAUUUGAUCGUUCGAAAUUCGAUCGUU----- ....................((.(((((.(((...........(((((((........)))))))............((((((.........))))))...))).))))).))..----- ( -17.90, z-score = 0.47, R) >consensus __________________AUCAUUAGGAAUGCCACGAUUUGGAAUU_CCAUGAUUAGGGAUUCCAGGAUUGGGAAUUCCAUGAUUGGGAAUGCCAGGAUUCGGGAUUCCAUCAUU_____ ........................................................((.(((((.......((....)).......))))).)).(((........)))........... ( -7.33 = -7.30 + -0.02)


| Location | 14,226,993 – 14,227,092 |
|---|---|
| Length | 99 |
| Sequences | 14 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 62.04 |
| Shannon entropy | 0.80540 |
| G+C content | 0.45725 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -6.92 |
| Energy contribution | -6.89 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 2.14 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.546749 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
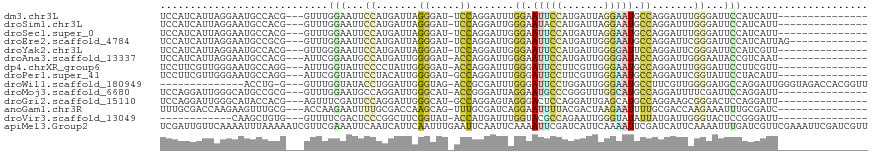
>dm3.chr3L 14226993 99 - 24543557 UCCAUCAUUAGGAAUGCCACG---GUUUGGAAUUCCAUGAUUAGGGAU-UCCAGGAUUUGGAAUUCCAUGAUUAGGAAUGCCAGGAUUUGGGAUUCCAUCAUU--------------- ..........(((((.(((..---.(((((.(((((.(((((((((((-(((((...)))))))))).))))))))))).)))))...))).)))))......--------------- ( -39.40, z-score = -3.52, R) >droSim1.chr3L 13582239 99 - 22553184 UCCAUCAUUAGGAAUGCCACG---GUUUGGAAUUCCAUGAUUAGGGAU-UCCAGGAUUGGGAAUACCAUGAUUAGGAAUGCCAGGAUUUGGGAUUCCAUCAUU--------------- ..........(((((.(((..---.(((((.(((((.((((((.((((-(((.......))))).)).))))))))))).)))))...))).)))))......--------------- ( -34.80, z-score = -2.23, R) >droSec1.super_0 6372845 99 - 21120651 UCCAUCAUUAGGAAUGCCACG---GUUUGGAAUUCCAUGAUUAGGGAU-UCCAGGAUUGGGAAUUCCAUGAUUAGGAAUGCCAGGAUUUGGGAUUCCAUCAUU--------------- ..........(((((.(((..---.(((((.(((((.(((((((((((-(((.......)))))))).))))))))))).)))))...))).)))))......--------------- ( -38.40, z-score = -2.99, R) >droEre2.scaffold_4784 14219169 101 - 25762168 UCCAUCAUUAGGAAUGCCGCG---GUUUGGAAUUCCAUGAUUAGGGAU-UCCAGGAUUGGGAAUUCCAUGAUUGGGAAUGCCAGGAUUCGGGAUUCCAUCAUUAG------------- ..........(((((.(((..---.(((((.(((((.(((((((((((-(((.......)))))))).))))))))))).)))))...))).)))))........------------- ( -38.10, z-score = -2.37, R) >droYak2.chr3L 14324561 99 - 24197627 UCCAUCAUUAGGAAUGCCACG---GUUGGGAAUUCCAUGAUUAGGGAU-UCCAGGAUUGGGAAUUCCAUGAUUGGGGAUUCCAGGAUUCGGGAUUCCAUCGUU--------------- ..........(((((.((...---....((((((((.......)))))-))).(((((.((((((((.......))))))))..))))))).)))))......--------------- ( -37.20, z-score = -1.58, R) >droAna3.scaffold_13337 18466010 99 + 23293914 UCCAUCAUUAGGAAUGCCACG---AUUCGGAAUGCCAUGAUUGGGGAU-ACCAGGAUUGGGAAUUCCAUGAUUGGGGAUACCAGGAUUGGGAAUACCGUCAAU--------------- ((((((((..(((((.((.((---((((((.((.((.......)).))-.)).)))))))).))))))))))(((.....))))))..((.....))......--------------- ( -28.60, z-score = -0.02, R) >dp4.chrXR_group6 6916498 99 - 13314419 UCCUUCGUUGGGAAUGCCAGG---AUUUGGUAUUCCCCUAUUGGGGAU-ACCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUUGGGAUUCCUUCGUU--------------- .....((..((((((.(((((---.((((((((.((((....))))))-)))))).(((((.((((((.....)))))).))))).))))).)))))).))..--------------- ( -44.10, z-score = -3.09, R) >droPer1.super_41 274448 99 - 728894 UCCUUCGUUGGGAAUGCCAGG---AUUCGGUAUUCCUACAUUGGGGAU-GCCAGGAUUUGGGAUUCCUUCGUUGGGAAUGCCAGGAUUCGGUAUUCCUACAUU--------------- ......((.(((((((((.((---((((((((((((((....((((((-.((((...)))).))))))....)))))))))).)))))))))))))))))...--------------- ( -46.60, z-score = -4.71, R) >droWil1.scaffold_180949 3514038 99 - 6375548 --------------ACCUG-G---GUUUGGUAUACCUGGAUUGGGUAG-ACCGCGAUUUGGGAUUCCUGGAUUGGGAAUGCCUUCGUUGGGGAUGCCAGGAUUGGGUAGACCACGGUU --------------(((..-(---.((((((((.((..(...(((((.-.((.(((((..((...))..)))))))..)))))...)..)).)))))))).)..))).(((....))) ( -40.30, z-score = -2.80, R) >droMoj3.scaffold_6680 8059378 99 + 24764193 UCCAGGAUUGGGCAUGCCGCG---GUUUGGAAUGCCAGGAUUGGGCAU-ACCGGGAUUAGGAAUGCCCGGGUUUGGCAUGCCAGGAUUUUCGAUUCCAGGAUU--------------- (((.......((((((((..(---((.......)))....((((((((-.((.......)).))))))))....)))))))).(((........))).)))..--------------- ( -40.60, z-score = -2.09, R) >droGri2.scaffold_15110 21437788 99 - 24565398 UCCAGGAUUGGGCAUACCACG---AGUUUCGAUUCCAGGAUUGGGCAU-GCCAGGAGUAGGGACUCCAGGAUUGAGCAUGCCAGGAAGCGGGACUCCAGGAUU--------------- (((.....(((.....))).(---(((..((.((((.......(((((-(((((((((....))))).....)).))))))).)))).))..))))..)))..--------------- ( -38.00, z-score = -2.54, R) >anoGam1.chr3R 30836173 99 + 53272125 UUUGCGACCAAGAAGUUUGCG---ACCAAGAAUUUUGCGACCAAGCAG-UUUGCGAUCAGGAAUUUUACGACUAAGAAUUUUGCGACCAAGAAAUUUGCGAUC--------------- ...((((.(..(((((((...---.....)))))))((......)).)-.))))((((.((((((((.......))))))))((((.........))))))))--------------- ( -14.90, z-score = 1.14, R) >droVir3.scaffold_13049 3121581 87 + 25233164 ------------CAAGCUGUG---GUUUUCGACUCCCGGCUUCGGUAU-ACCAUGAUUUGGUACGCCAGAAUUGGGUAUAUUAUGAUUGGGUACUCCGGGAUU--------------- ------------.((((((.(---(.........))))))))(((..(-(((.((((((((((..((.......))..))))).)))))))))..))).....--------------- ( -23.50, z-score = -0.15, R) >apiMel3.Group2 14092252 118 + 14465785 UCGAUUGUUCAAAAUUUAAAAAUCGUUCGAAAUUCAAUCAUUCAAUUUGAAUUCAAUUCAAAAUUCGAUCAUUCAAAAUUCGAUCAUUCAAAAUUUGAUCGUUCGAAAUUCGAUCGUU .(((((..............))))).(((((.(((...........(((((((........)))))))............((((((.........))))))...))).)))))..... ( -18.14, z-score = 1.31, R) >consensus UCCAUCAUUAGGAAUGCCACG___GUUUGGAAUUCCAUGAUUAGGGAU_ACCAGGAUUGGGAAUUCCAUGAUUGGGAAUGCCAGGAUUCGGGAUUCCAUCAUU_______________ ............................(((...((.......((.....)).......((.(((((.......))))).)).......))...)))..................... ( -6.92 = -6.89 + -0.03)
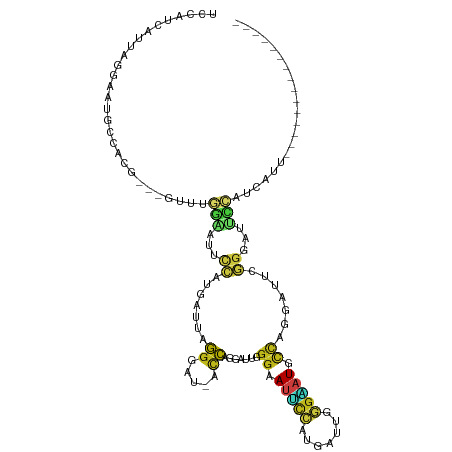
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:21 2011