| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,200,698 – 14,200,802 |
| Length | 104 |
| Max. P | 0.608406 |

| Location | 14,200,698 – 14,200,802 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 69.71 |
| Shannon entropy | 0.61912 |
| G+C content | 0.34367 |
| Mean single sequence MFE | -21.28 |
| Consensus MFE | -8.36 |
| Energy contribution | -8.04 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.608406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
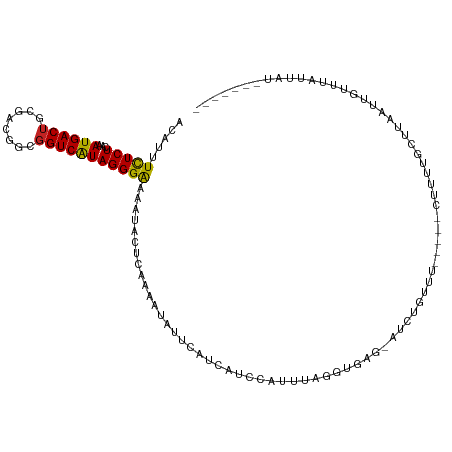
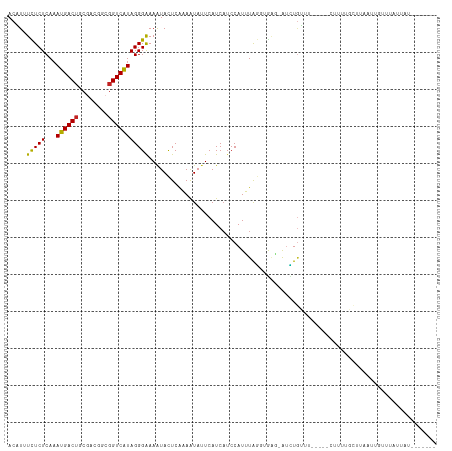
>dm3.chr3L 14200698 104 - 24543557 ACAUGUCUCUCAAAUGACUGCGACGGCGGUCAUAGGGAAAAUACUCAAAAUAUUCAUCAUCCAUUUAGGUGAG-AUCUGUUUU----CGUUAACUGAAUUGUUUAUUUA------- .....(((((...((((((((....)))))))))))))(((((..(((.....((.(((((......))))))-)((.(((..----....))).)).)))..))))).------- ( -23.10, z-score = -1.45, R) >droSim1.chr3L 13556271 104 - 22553184 ACAUGUCUCUCAAAUGACUGCGACGGCGGUCAUAGGGAAAAUACUCAAAAUAUUCAUCAUCCAUUUAGGUGAG-AUCUGUUUU----CUUUAACUUAAUUGUUUAUUUG------- .....(((((...((((((((....)))))))))))))(((((..(((.....((.(((((......))))))-)...(((..----....)))....)))..))))).------- ( -21.70, z-score = -1.65, R) >droSec1.super_0 6347304 104 - 21120651 ACAUGUCUCUCAAAUGACUGCGACGGCGGUCAUAGGGAAAAUACUCAAAAUAUUCAUCAUCCAUUUAGGUGAG-AUCUGUUUU----CUUUAACUUAAUUGUUUAUUUG------- .....(((((...((((((((....)))))))))))))(((((..(((.....((.(((((......))))))-)...(((..----....)))....)))..))))).------- ( -21.70, z-score = -1.65, R) >droYak2.chr3L 14298156 108 - 24197627 ACAUGUCUCUCAAAUGACUGCGACGGCGGUCAUAGGGAAAAUACUCAAAAUAUUCAUCAUCCAUUUAGUUGAG-AUCUAUGUAUUUGCUUUUGCUUAAUUGUUUAUGAA------- (((((..((((((((((((((....)))))))).(((......)))......................)))))-)..)))))....((....))...............------- ( -23.60, z-score = -1.42, R) >droEre2.scaffold_4784 14192941 104 - 25762168 ACAUGUCUCUCAAAUGACUGCGACGGCGGUCGUAGGGGAAAUACUCAAAAUAUUCAUCUUCCAUUUAGGUGGA-AUCUGU-UAU---CUUUUGCUUAAUUGUUUAUUAU------- ...(((.((((..((((((((....))))))))..)))).)))..(((((.((.((..((((((....)))))-)..)).-.))---.)))))................------- ( -24.00, z-score = -2.02, R) >droAna3.scaffold_13337 18440994 102 + 23293914 ACUUUUCUCUCAAAUGACUGCGACGACGGUCAUAGGGAAAAUACUCGAAAUACUCAUCAUCCAUUUAG-CAAG-AUUUGAU------GUUUUAUUUUUAUGUUUUUUAAU------ ..((((((((...(((((((......))))))))))))))).....((((((..(((((...((((..-..))-)).))))------)...)))))).............------ ( -19.10, z-score = -1.46, R) >droPer1.super_41 249004 89 - 728894 ACUUUUUUCUCAAAUGACUGCGACGACGGUCAUAGGGAAAGUACUCAAAAUACUCAUCAUCCAUUUAUGUGAGAAUGUGAG------AUUCCGCG--------------------- ....(((((((..(((((((......))))))).)))))))..((((.....((((.(((......)))))))....))))------........--------------------- ( -21.00, z-score = -1.30, R) >dp4.chrXR_group6 6890210 89 - 13314419 ACUUUUUUCUCAAAUGACUGCGACGACGGUCAUAGGGAAAGUACUCAAAAUACUCAUCAUCCAUUUAUGUGAGAAUGUGAG------AUUCGGCG--------------------- ....(((((((..(((((((......))))))).)))))))..((((.....((((.(((......)))))))....))))------........--------------------- ( -21.00, z-score = -1.15, R) >droMoj3.scaffold_6680 8022800 100 + 24764193 ACGUUUUUCUCAUAUGACUGC------GGUCAUAGGGAAAAUAUUCAUAAUAUUCAUCAUCCAUAUGA--A---GUUUGUUG-----CCUCUGUAGAUUUAUAUAGAGUAUAUAUA ....(((((((.((((((...------.)))))))))))))......(((((..(.((((....))))--.---)..)))))-----.(((((((......)))))))........ ( -23.00, z-score = -2.00, R) >droVir3.scaffold_13049 3088466 99 + 25233164 ACAUUUUUCUCAAAUGACUGC------GGUCAUAGGGAAAAUAUUCAUAAUAUUCAUCAUCCAUAUGA-GA---GAUUGUUG------CUCUGGAGUUUUGUA-AUUGUGUAUAAA ....(((((((..(((((...------.))))).)))))))(((.((((((...((...((((...((-(.---........------)))))))....))..-)))))).))).. ( -19.20, z-score = -0.34, R) >droGri2.scaffold_15110 21405446 104 - 24565398 ACAUUUUUCUCAAAUGACGGC------GGUCAUAGGGAAAAUAUUCAUAGUAUUCAUCAUCCAUAUGA-AGAUUGCUUCUUG-----GUUUUAUUUAUUUUUGUUUUGUUUUUGUA (((.(((((((..(((((...------.))))).)))))))...(((.((((.((.((((....))))-.)).))))...))-----)........................))). ( -16.70, z-score = -0.55, R) >consensus ACAUUUCUCUCAAAUGACUGCGACGGCGGUCAUAGGGAAAAUACUCAAAAUAUUCAUCAUCCAUUUAGGUGAG_AUCUGUUU_____CUUUUGCUUAAUUGUUUAUUAU_______ .....(((((...((((((........))))))))))).............................................................................. ( -8.36 = -8.04 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:16 2011