
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,197,537 – 14,197,638 |
| Length | 101 |
| Max. P | 0.980774 |

| Location | 14,197,537 – 14,197,638 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 71.67 |
| Shannon entropy | 0.53021 |
| G+C content | 0.52822 |
| Mean single sequence MFE | -36.76 |
| Consensus MFE | -24.82 |
| Energy contribution | -26.26 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
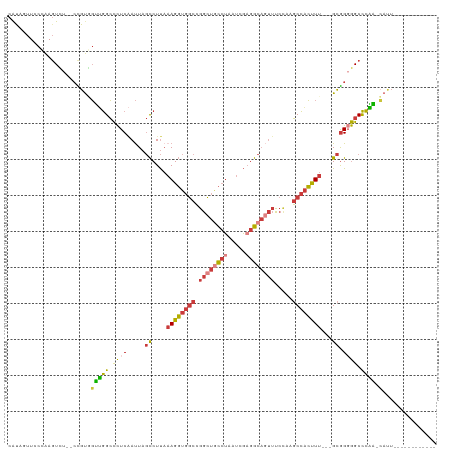
>dm3.chr3L 14197537 101 + 24543557 CAUAGUUCCCAAGUCU--ACGUGGUUGGCCCUCAAUUAGCCUUAAAGGUGGCCGGCUGCCUAAUCGAGGCAGUUUCCAAGCCACUUU---GCGGGGGCCCAA-CAUU------------ ................--.....((((((((((.....((....((((((((.((((((((.....)))))))).....))))))))---))))))).))))-)...------------ ( -36.20, z-score = -1.37, R) >droSim1.chr3L 13553218 102 + 22553184 CAUAGUUCCCAAGUCU--CCGUGGUUGGCCCUCAAUUAGCCUUAAAGGUGGCCGGCUGCCUAAUCGAGGCAGUUUCCAAGCCACUUU---GCGGGGGCCCAAACAUU------------ ............((((--((((((((((.......))))))...((((((((.((((((((.....)))))))).....))))))))---)))))))).........------------ ( -37.30, z-score = -1.67, R) >droSec1.super_0 6344296 102 + 21120651 CAUAGUUCCCAAGUCU--CCGUGGUUGGCCCUCAAUUAGCCUUAAAGGUGGCCGGCUGCCUAAUCGAGGCAGUUUCCAAGCCACUUU---GCGGGGGCCCAAACAUU------------ ............((((--((((((((((.......))))))...((((((((.((((((((.....)))))))).....))))))))---)))))))).........------------ ( -37.30, z-score = -1.67, R) >droYak2.chr3L 14293356 113 + 24197627 CACUAUUCCCAAGUCU--CCGUGGUUGGCCCUCAAUUAGCCUUAAAGGUGGCCGGCUGCCUAAUCGAGGCAGUUUCCAAGCCACUUU---GCGGGGGCCCAG-CAUUGUUGGCAGUUUG .(((........((((--((((((((((.......))))))...((((((((.((((((((.....)))))))).....))))))))---))))))))((((-(...))))).)))... ( -40.90, z-score = -1.04, R) >droEre2.scaffold_4784 14189814 101 + 25762168 CACAGUUCCCAAGUCU--CCGCUGUUGGCCCUCAAUUAGCCUUAAAGGUGGCCGGCUGCCUAAUCGAGGCAGUUUCCAAGCCACUUU---GCGGGGGCCCAA-CAUU------------ ............((((--((((....(((.........)))...((((((((.((((((((.....)))))))).....))))))))---))))))))....-....------------ ( -38.20, z-score = -2.07, R) >droAna3.scaffold_13337 18438343 108 - 23293914 CAUAAUUACAGAGUGUAGCUGUUGGGGGCCCUCGAUUAGCCUUUUAGGUGGCCGGCUACCUAAUGGCGGCAUUUUCUGAGCCACCUUUAAGCGGCGGCCCCC-AACUAA---------- .....((((.....))))..((((((((((.(((.((((((..((((((((....)))))))).)))(((.........))).....))).))).)))))))-)))...---------- ( -46.80, z-score = -2.95, R) >droVir3.scaffold_13049 3085653 101 - 25233164 -GUCAAUCCCAA-UUUGGCUGCAUUUAGAUUUCCACU--UCUCAAAGCGCACAAAUUAUAGAGUUUAGGCUCGAUC-AAUCUGGCUUAAGGUAGCAGCUUGAUAAA------------- -.........((-((((((.((.((.(((........--))).)).)))).))))))...((((....)))).(((-((.(((.((......))))).)))))...------------- ( -20.60, z-score = -0.16, R) >consensus CAUAGUUCCCAAGUCU__CCGUGGUUGGCCCUCAAUUAGCCUUAAAGGUGGCCGGCUGCCUAAUCGAGGCAGUUUCCAAGCCACUUU___GCGGGGGCCCAA_CAUU____________ ....................(((.(((((((((.....((....((((((((.((((((((.....)))))))).....))))))))...)))))))).))).)))............. (-24.82 = -26.26 + 1.44)


| Location | 14,197,537 – 14,197,638 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 71.67 |
| Shannon entropy | 0.53021 |
| G+C content | 0.52822 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -19.25 |
| Energy contribution | -20.21 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
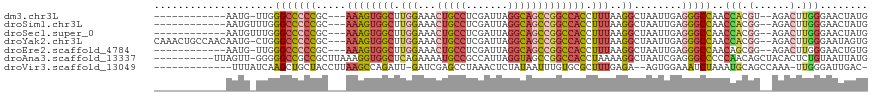
>dm3.chr3L 14197537 101 - 24543557 ------------AAUG-UUGGGCCCCCGC---AAAGUGGCUUGGAAACUGCCUCGAUUAGGCAGCCGGCCACCUUUAAGGCUAAUUGAGGGCCAACCACGU--AGACUUGGGAACUAUG ------------...(-((.(((((((..---(((((((((.((...((((((.....))))))))))))).))))..))........)))))..(((.(.--...).))).))).... ( -33.60, z-score = -0.45, R) >droSim1.chr3L 13553218 102 - 22553184 ------------AAUGUUUGGGCCCCCGC---AAAGUGGCUUGGAAACUGCCUCGAUUAGGCAGCCGGCCACCUUUAAGGCUAAUUGAGGGCCAACCACGG--AGACUUGGGAACUAUG ------------...((((.(((((((..---(((((((((.((...((((((.....))))))))))))).))))..))........)))))..(((.(.--...).))))))).... ( -38.40, z-score = -1.66, R) >droSec1.super_0 6344296 102 - 21120651 ------------AAUGUUUGGGCCCCCGC---AAAGUGGCUUGGAAACUGCCUCGAUUAGGCAGCCGGCCACCUUUAAGGCUAAUUGAGGGCCAACCACGG--AGACUUGGGAACUAUG ------------...((((.(((((((..---(((((((((.((...((((((.....))))))))))))).))))..))........)))))..(((.(.--...).))))))).... ( -38.40, z-score = -1.66, R) >droYak2.chr3L 14293356 113 - 24197627 CAAACUGCCAACAAUG-CUGGGCCCCCGC---AAAGUGGCUUGGAAACUGCCUCGAUUAGGCAGCCGGCCACCUUUAAGGCUAAUUGAGGGCCAACCACGG--AGACUUGGGAAUAGUG ...............(-((((((((((..---(((((((((.((...((((((.....))))))))))))).))))..))........)))))..(((.(.--...).)))...)))). ( -38.80, z-score = -0.71, R) >droEre2.scaffold_4784 14189814 101 - 25762168 ------------AAUG-UUGGGCCCCCGC---AAAGUGGCUUGGAAACUGCCUCGAUUAGGCAGCCGGCCACCUUUAAGGCUAAUUGAGGGCCAACAGCGG--AGACUUGGGAACUGUG ------------..(.-(..((.(.((((---...((((((.((...((((((.....))))))))))))))......((((.......))))....))))--.).))..).)...... ( -36.30, z-score = -0.56, R) >droAna3.scaffold_13337 18438343 108 + 23293914 ----------UUAGUU-GGGGGCCGCCGCUUAAAGGUGGCUCAGAAAAUGCCGCCAUUAGGUAGCCGGCCACCUAAAAGGCUAAUCGAGGGCCCCCAACAGCUACACUCUGUAAUUAUG ----------...(((-(((((((((..(((((.((((((.........)))))).)))))..)).((((........)))).......))))))))))...(((.....)))...... ( -49.10, z-score = -4.05, R) >droVir3.scaffold_13049 3085653 101 + 25233164 -------------UUUAUCAAGCUGCUACCUUAAGCCAGAUU-GAUCGAGCCUAAACUCUAUAAUUUGUGCGCUUUGAGA--AGUGGAAAUCUAAAUGCAGCCAAA-UUGGGAUUGAC- -------------........(((((...(((((((((((((-(...(((......)))..))))))).))...))))).--..(((....)))...)))))....-...........- ( -18.80, z-score = 0.48, R) >consensus ____________AAUG_UUGGGCCCCCGC___AAAGUGGCUUGGAAACUGCCUCGAUUAGGCAGCCGGCCACCUUUAAGGCUAAUUGAGGGCCAACCACGG__AGACUUGGGAACUAUG ....................(((((.(((...((((((((.(((...(((((.......))))))))))))).)))...)).....).))))).......................... (-19.25 = -20.21 + 0.97)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:15 2011