| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,170,856 – 14,170,958 |
| Length | 102 |
| Max. P | 0.931536 |

| Location | 14,170,856 – 14,170,951 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.38 |
| Shannon entropy | 0.47391 |
| G+C content | 0.44185 |
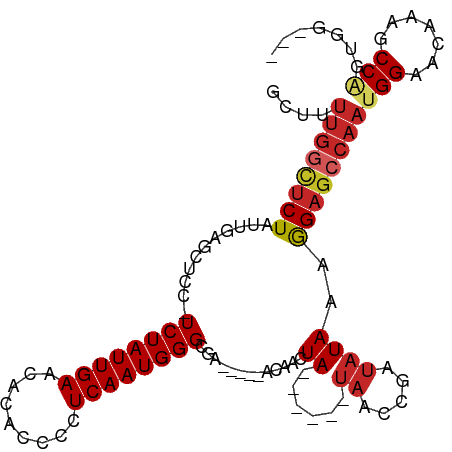
| Mean single sequence MFE | -21.34 |
| Consensus MFE | -10.86 |
| Energy contribution | -11.02 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
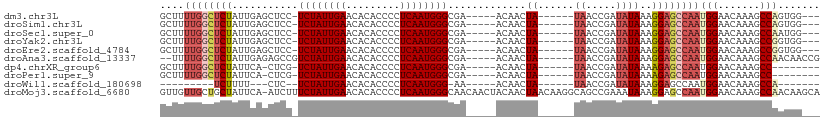
>dm3.chr3L 14170856 95 - 24543557 ---GCUCUAUUG-AGCUCCUCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCAGUGGAACAAUG--------- ---(.(((((((-.(((..(((((((......((((.....))).).-----.......------...((........))...)))))))....)))))))))).)....--------- ( -22.80, z-score = -2.29, R) >droSim1.chr3L 13526964 95 - 22553184 ---GCUCUAUUG-AGCUCCUCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCAGUGGAACAAUG--------- ---(.(((((((-.(((..(((((((......((((.....))).).-----.......------...((........))...)))))))....)))))))))).)....--------- ( -22.80, z-score = -2.29, R) >droSec1.super_0 6317934 95 - 21120651 ---GCUCUAUUG-AGCUCCUCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCAAUGGAACAAUG--------- ---(.(((((((-.(((..(((((((......((((.....))).).-----.......------...((........))...)))))))....)))))))))).)....--------- ( -22.80, z-score = -2.92, R) >droYak2.chr3L 14265695 95 - 24197627 ---GCUCUAUUG-AGCUCCUCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCGGUGGAACAAAG--------- ---(.(((((((-.(((((((((((((.........)))))))....-----.....((------((.....)))).))))))))))))).)..................--------- ( -22.20, z-score = -1.80, R) >droEre2.scaffold_4784 14161693 95 - 25762168 ---GCUCUAUUG-AGCUCCUCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCGGUGGAACAAUG--------- ---(.(((((((-.(((((((((((((.........)))))))....-----.....((------((.....)))).))))))))))))).)..................--------- ( -22.20, z-score = -1.78, R) >droAna3.scaffold_13337 18415902 105 + 23293914 ---GCUCUAUUGAGAGCCGUCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCAACAACCGGGACCAAACAAUG ---(((((....)))))((((((((((.........)))))))))).-----.......------...(((.......(....)..(((.......))).....)))............ ( -22.20, z-score = -1.74, R) >dp4.chrXR_group6 4859314 89 - 13314419 ---GCUCUAUU--CACUCGUCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAAGAGCCAAUGGAACAAAGCCAACAAUA-------------- ---(((((...--...(((((((((((.........)))))))))))-----.....((------((.....))))..)))))...(((.......)))......-------------- ( -19.80, z-score = -3.94, R) >droPer1.super_9 3161455 89 - 3637205 ---GCUCUAUU--CACUCGUCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAAGAGCCAAUGGAACAAAGCCAACAAUA-------------- ---(((((...--...(((((((((((.........)))))))))))-----.....((------((.....))))..)))))...(((.......)))......-------------- ( -19.80, z-score = -3.94, R) >droWil1.scaffold_180698 10072477 78 + 11422946 ----------------CUCUCUAUUGAACACACCCCUCAAUGGG-AA-----ACAACUA------UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCAAACAAUA------------- ----------------...(((((((.......(((.....)))-..-----.......------...((........))...)))))))................------------- ( -12.70, z-score = -1.72, R) >droVir3.scaffold_13049 18444054 116 - 25233164 CUGCUGCUAUUC-AGUCUUUCUAUUGAACACACCCCUCAAUGGGCAACAACUACAACUAACAAGGCGGCCGAAAUAAAGGAGCCAAUGGAACAAAGCCAACAAGCAGCGUAACAAUG-- ..((((((....-.((..(((((((........(((.....)))...................(((..((........)).))))))))))....)).....)))))).........-- ( -25.80, z-score = -1.09, R) >droMoj3.scaffold_6680 19241869 113 - 24764193 ---CUGCUAUUC-AAUCUUUCUAUUGAACACACCCCUCAAUGGGCAACAACUACAACUAACAAGGCAGCCGAAAUAAAGGAGCCAAUGGAACAAAGCCAACAAGCAGCGUAACAAUA-- ---(((((.(((-(((......)))))).....(((.....)))...................(((..((........)).)))..(((.......)))...)))))..........-- ( -19.00, z-score = -0.28, R) >droGri2.scaffold_15110 15359890 113 + 24565398 ---CUGCUAUUC-AGUGUUUCUAUUGAACACACCCCUCAAUGGGCAACAACUACAACUAACAAGGCGGCCGAUAUAAAGGAGCCAAUGGAACAAAGCCAACAAGCAGCGUAACAAUG-- ---(((((....-((((((((((((((.........)))))))).))).)))...........(((..((........)).)))..(((.......)))...)))))..........-- ( -24.00, z-score = -0.58, R) >consensus ___GCUCUAUUG_AGCUCCUCUAUUGAACACACCCCUCAAUGGGCGA_____ACAACUA______UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCAACAAAACAAUG_________ ...................(((((((.......(((.....)))........................((........))...)))))))............................. (-10.86 = -11.02 + 0.17)
| Location | 14,170,863 – 14,170,958 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 85.39 |
| Shannon entropy | 0.29382 |
| G+C content | 0.44901 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
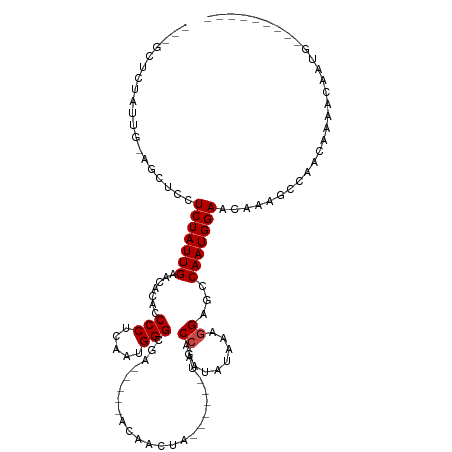
>dm3.chr3L 14170863 95 - 24543557 GCUUUUGGCUCUAUUGAGCUCC-UCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCAGUGG--- .((.((((((((((((.(((((-((((((((.........)))))))....-----.....((------((.....)))).))))))))))))).....))))).))--- ( -27.10, z-score = -2.76, R) >droSim1.chr3L 13526971 95 - 22553184 GCUUUUGGCUCUAUUGAGCUCC-UCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCAGUGG--- .((.((((((((((((.(((((-((((((((.........)))))))....-----.....((------((.....)))).))))))))))))).....))))).))--- ( -27.10, z-score = -2.76, R) >droSec1.super_0 6317941 95 - 21120651 GCUUUUGGCUCUAUUGAGCUCC-UCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCAAUGG--- .((.((((((((((((.(((((-((((((((.........)))))))....-----.....((------((.....)))).))))))))))))).....))))).))--- ( -27.40, z-score = -3.42, R) >droYak2.chr3L 14265702 95 - 24197627 GCUUUUGGCUCUAUUGAGCUCC-UCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCGGUGG--- (((((.(..(((((((.(((((-((((((((.........)))))))....-----.....((------((.....)))).))))))))))))).))))))......--- ( -26.60, z-score = -2.32, R) >droEre2.scaffold_4784 14161700 95 - 25762168 GCUUUUGGCUCUAUUGAGCUCC-UCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCGGUGG--- (((((.(..(((((((.(((((-((((((((.........)))))))....-----.....((------((.....)))).))))))))))))).))))))......--- ( -26.60, z-score = -2.32, R) >droAna3.scaffold_13337 18415915 97 + 23293914 --UUUUGGCUCUAUUGAGAGCCGUCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCAACAACCG --..((((((((((((.(..(((((((((((.........)))))))))..-----.....((------((.....))))..))..)))))))).....)))))...... ( -24.90, z-score = -3.11, R) >dp4.chrXR_group6 4859321 89 - 13314419 GCUUUUGGCUCUAUUCA-CUCG-UCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAAGAGCCAAUGGAACAAAGCC-------- .((.((((((((.....-.(((-((((((((.........)))))))))))-----.....((------((.....))))..)))))))).)).........-------- ( -25.60, z-score = -4.75, R) >droPer1.super_9 3161462 89 - 3637205 GCUUUUGGCUCUAUUCA-CUCG-UCUAUUGAACACACCCCUCAAUGGGCGA-----ACAACUA------UAACCGAUAUAAAAGAGCCAAUGGAACAAAGCC-------- .((.((((((((.....-.(((-((((((((.........)))))))))))-----.....((------((.....))))..)))))))).)).........-------- ( -25.60, z-score = -4.75, R) >droWil1.scaffold_180698 10072484 77 + 11422946 ---------UCUUUU---CUC--UCUAUUGAACACACCCCUCAAUGGG-AA-----ACAACUA------UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCA------- ---------......---...--(((((((.......(((.....)))-..-----.......------...((........))...))))))).........------- ( -12.70, z-score = -1.06, R) >droMoj3.scaffold_6680 19241880 109 - 24764193 GUUGUUGCUGCUAUUCA-AUCUUUCUAUUGAACACACCCCUCAAUGGGCAACAACUACAACUAACAAGGCAGCCGAAAUAAAGGAGCCAAUGGAACAAAGCCAACAAGCA ((((((((.....((((-((......))))))......((.....))))))))))............(((..((........)).)))..(((.......)))....... ( -23.00, z-score = -0.84, R) >consensus GCUUUUGGCUCUAUUGAGCUCC_UCUAUUGAACACACCCCUCAAUGGGCGA_____ACAACUA______UAACCGAUAUAAAGGAGCCAAUGGAACAAAGCCAGUGG___ ....((((((((...........((....))......(((.....)))..................................))))))))(((.......)))....... (-12.86 = -13.40 + 0.54)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:12 2011