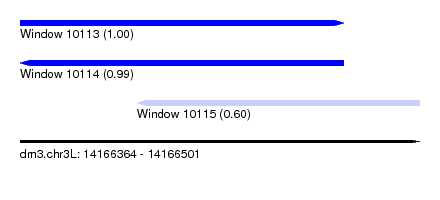
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,166,364 – 14,166,501 |
| Length | 137 |
| Max. P | 0.996488 |

| Location | 14,166,364 – 14,166,475 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.18 |
| Shannon entropy | 0.42334 |
| G+C content | 0.44035 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -16.21 |
| Energy contribution | -16.82 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.996488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
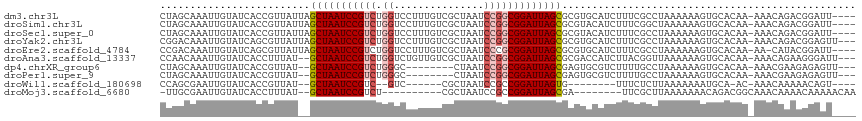
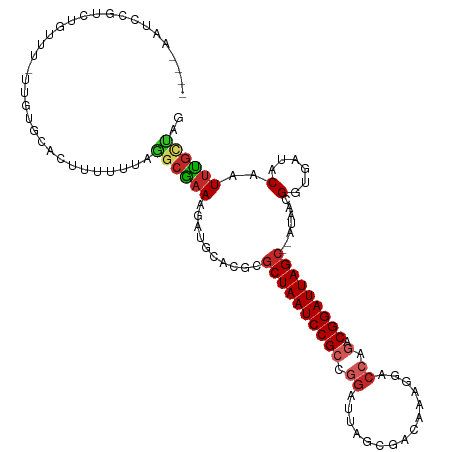
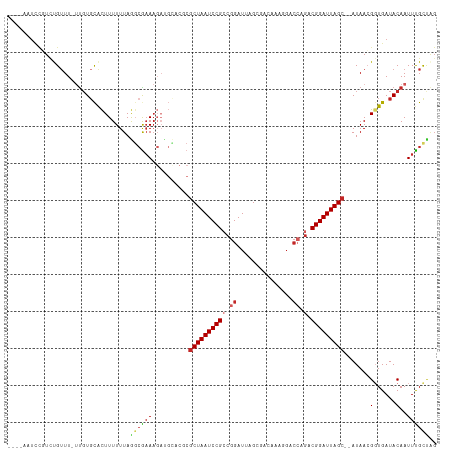
>dm3.chr3L 14166364 111 + 24543557 CUAGCAAAUUGUAUCACCGUUAUUAGCUAAUCCGUCUGGUCCUUUGUCGCUAAUCCGGCGGAUUAGCGCGUGCAUCUUUCGCCUAAAAAAGUGCACAA-AAACAGACGGAUU---- .((((.(((............))).))))(((((((((.......(.(((((((((...))))))))))(((((.((((........)))))))))..-...))))))))).---- ( -38.50, z-score = -4.65, R) >droSim1.chr3L 13522444 111 + 22553184 CUAGCAAAUUGUAUCACCGUUAUUAGCUAAUCCGUCUGGUCCUUUGUCGCUAAUCCGGCGGAUUAGCGCGUACAUCUUUCGGCUAAAAAAGUGCACAA-AAACAGACGGAUU---- .((((.(((............))).))))(((((((((....((((((((((((((...))))))))).((((.................))))))))-)..))))))))).---- ( -32.73, z-score = -2.89, R) >droSec1.super_0 6313419 111 + 21120651 CUAGCAAAUUGUAUCACCGUUAUUAGCUAAUCCGUCUGGUCCUUUGUCGCUAAUCCGGCGGAUUAGCGCGUACAUCUUUCGCCUAAAAAAGUGCACAA-AAACAGACGGAUU---- .((((.(((............))).))))(((((((((....((((((((((((((...))))))))).((((.................))))))))-)..))))))))).---- ( -32.73, z-score = -3.23, R) >droYak2.chr3L 14260931 112 + 24197627 CGGACAAAUUGUAUCAGCGUUAUUAGCUAAUCCGUCUGGUCCUUUGUCGCUAAUCCGGCGGAUUAGCGCGUGCAUCUUUCGCCUAAAAAAGUGCACAA-AAACAGACGGAGUU--- ..(((..........(((.......)))..((((((((.......(.(((((((((...))))))))))(((((.((((........)))))))))..-...)))))))))))--- ( -38.90, z-score = -3.73, R) >droEre2.scaffold_4784 14157148 110 + 25762168 CCGACAAAUUGUAUCAGCGUUAUUAGCUAAUCCGUCUGGUCCUUUGUCGCUAAUCCCGCGGAUUAGCGCGUGCAUCUUUCGCCUAAAAAAGUGCACAA-AA-CAUACGGAUU---- ...............(((.......)))(((((((.((.......(.(((((((((...))))))))))(((((.((((........)))))))))..-..-)).)))))))---- ( -33.10, z-score = -3.20, R) >droAna3.scaffold_13337 18411632 110 - 23293914 CCAACAAAUUGUAUCACCUUUAU--GCUAAUCCGUCUGGUCUGUUGUCGCUAAUCCGGCGGAUUAGCGCGACCAUCUUACGGUUAAAAAAGUGCACAA-AAACAGAAGGGAUU--- ............(((.(((((..--(((((((((((.(((..((....))..))).)))))))))))..((((.......))))..............-.....)))))))).--- ( -30.60, z-score = -2.30, R) >dp4.chrXR_group6 4854335 102 + 13314419 CUAGCAAAUUGUAUCACCGUUAU--GCUAAUCCGUCUGGGC--------CUAAUCCGGCGGAUUAGCGAGUGCGUCUUUUGCCUAAAAAAGUGCACAA-AAACGAAGAGAGUU--- .............((..((((.(--(((((((((.(((((.--------....))))))))))))))).(((((.(((((......))))))))))..-.))))..)).....--- ( -35.10, z-score = -3.83, R) >droPer1.super_9 3156468 102 + 3637205 CUAGCAAAUUGUAUCACCGUUAU--GCUAAUCCGUCUGGGC--------CUAAUCCGGCGGAUUAGCGAGUGCGUCUUUUGCCUAAAAAAGUGCACAA-AAACGAAGAGAGUU--- .............((..((((.(--(((((((((.(((((.--------....))))))))))))))).(((((.(((((......))))))))))..-.))))..)).....--- ( -35.10, z-score = -3.83, R) >droWil1.scaffold_180698 10066946 92 - 11422946 CCAGCGAAUUGUAUCACCGUUAU--GCUAAUCCGUC--GUC------CGCUAAUCCGCCGGAUUAGUG--------UUUCUCUUAAAAAAAUGCA-AC-AAACAAAAACAGU---- ...((((......))...(..((--(((((((((.(--(..------........)).))))))))))--------)..)............)).-..-.............---- ( -16.50, z-score = -1.67, R) >droMoj3.scaffold_6680 19236935 95 + 24764193 -UUGCGAAUUGUAUCACCUUUAU--GCUAAUCCGUCU----------CGCUAAUCCGCCGGAUUAGCGA--------UUCGCUUAAAAAAACAGACGGCAAACAAAACAAAAACAA -..((((((((((.......)))--(((((((((...----------.((......)))))))))))))--------))))).................................. ( -21.30, z-score = -2.54, R) >consensus CUAGCAAAUUGUAUCACCGUUAU__GCUAAUCCGUCUGGUCCUUUGUCGCUAAUCCGGCGGAUUAGCGCGUGCAUCUUUCGCCUAAAAAAGUGCACAA_AAACAGACGGAGU____ .........................(((((((((.((((...............)))))))))))))................................................. (-16.21 = -16.82 + 0.61)

| Location | 14,166,364 – 14,166,475 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.18 |
| Shannon entropy | 0.42334 |
| G+C content | 0.44035 |
| Mean single sequence MFE | -34.21 |
| Consensus MFE | -14.43 |
| Energy contribution | -14.81 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14166364 111 - 24543557 ----AAUCCGUCUGUUU-UUGUGCACUUUUUUAGGCGAAAGAUGCACGCGCUAAUCCGCCGGAUUAGCGACAAAGGACCAGACGGAUUAGCUAAUAACGGUGAUACAAUUUGCUAG ----(((((((((((((-((((((((((((......))))).)))))((((((((((...))))))))).).))))).))))))))))..........((..(......)..)).. ( -41.50, z-score = -4.97, R) >droSim1.chr3L 13522444 111 - 22553184 ----AAUCCGUCUGUUU-UUGUGCACUUUUUUAGCCGAAAGAUGUACGCGCUAAUCCGCCGGAUUAGCGACAAAGGACCAGACGGAUUAGCUAAUAACGGUGAUACAAUUUGCUAG ----(((((((((((((-((((((((((((......))))).)))))((((((((((...))))))))).).))))).))))))))))..........((..(......)..)).. ( -39.50, z-score = -4.70, R) >droSec1.super_0 6313419 111 - 21120651 ----AAUCCGUCUGUUU-UUGUGCACUUUUUUAGGCGAAAGAUGUACGCGCUAAUCCGCCGGAUUAGCGACAAAGGACCAGACGGAUUAGCUAAUAACGGUGAUACAAUUUGCUAG ----(((((((((((((-((((((((((((......))))).)))))((((((((((...))))))))).).))))).))))))))))..........((..(......)..)).. ( -39.50, z-score = -4.43, R) >droYak2.chr3L 14260931 112 - 24197627 ---AACUCCGUCUGUUU-UUGUGCACUUUUUUAGGCGAAAGAUGCACGCGCUAAUCCGCCGGAUUAGCGACAAAGGACCAGACGGAUUAGCUAAUAACGCUGAUACAAUUUGUCCG ---...(((((((((((-((((((((((((......))))).)))))((((((((((...))))))))).).))))).))))))))(((((.......)))))............. ( -41.00, z-score = -4.84, R) >droEre2.scaffold_4784 14157148 110 - 25762168 ----AAUCCGUAUG-UU-UUGUGCACUUUUUUAGGCGAAAGAUGCACGCGCUAAUCCGCGGGAUUAGCGACAAAGGACCAGACGGAUUAGCUAAUAACGCUGAUACAAUUUGUCGG ----(((((((..(-((-((((((((((((......))))).)))))((((((((((...))))))))).)..)))))...)))))))...........((((((.....)))))) ( -36.80, z-score = -3.31, R) >droAna3.scaffold_13337 18411632 110 + 23293914 ---AAUCCCUUCUGUUU-UUGUGCACUUUUUUAACCGUAAGAUGGUCGCGCUAAUCCGCCGGAUUAGCGACAACAGACCAGACGGAUUAGC--AUAAAGGUGAUACAAUUUGUUGG ---....((........-((((((((((((((((((((....(((((.(((((((((...)))))))))......))))).)))).)))).--..))))))).)))))......)) ( -35.24, z-score = -3.51, R) >dp4.chrXR_group6 4854335 102 - 13314419 ---AACUCUCUUCGUUU-UUGUGCACUUUUUUAGGCAAAAGACGCACUCGCUAAUCCGCCGGAUUAG--------GCCCAGACGGAUUAGC--AUAACGGUGAUACAAUUUGCUAG ---.........(((((-((.(((.((.....)))))))))))).....((((((((((.((.....--------..)).).)))))))))--.....((..(......)..)).. ( -27.40, z-score = -1.81, R) >droPer1.super_9 3156468 102 - 3637205 ---AACUCUCUUCGUUU-UUGUGCACUUUUUUAGGCAAAAGACGCACUCGCUAAUCCGCCGGAUUAG--------GCCCAGACGGAUUAGC--AUAACGGUGAUACAAUUUGCUAG ---.........(((((-((.(((.((.....)))))))))))).....((((((((((.((.....--------..)).).)))))))))--.....((..(......)..)).. ( -27.40, z-score = -1.81, R) >droWil1.scaffold_180698 10066946 92 + 11422946 ----ACUGUUUUUGUUU-GU-UGCAUUUUUUUAAGAGAAA--------CACUAAUCCGGCGGAUUAGCG------GAC--GACGGAUUAGC--AUAACGGUGAUACAAUUCGCUGG ----..((((...((((-((-(((.(((((....))))).--------(.(((((((...))))))).)------).)--))))))).)))--)...((((((......)))))). ( -23.50, z-score = -1.98, R) >droMoj3.scaffold_6680 19236935 95 - 24764193 UUGUUUUUGUUUUGUUUGCCGUCUGUUUUUUUAAGCGAA--------UCGCUAAUCCGGCGGAUUAGCG----------AGACGGAUUAGC--AUAAAGGUGAUACAAUUCGCAA- ............((((..((((((((((....)))))..--------((((((((((...)))))))))----------))))))...)))--).....((((......))))..- ( -30.30, z-score = -2.93, R) >consensus ____AAUCCGUCUGUUU_UUGUGCACUUUUUUAGGCGAAAGAUGCACGCGCUAAUCCGCCGGAUUAGCGACAAAGGACCAGACGGAUUAGC__AUAACGGUGAUACAAUUUGCUAG .................................((((((..........((((((((((.((...............)).).))))))))).......(......)..)))))).. (-14.43 = -14.81 + 0.38)
| Location | 14,166,404 – 14,166,501 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 79.34 |
| Shannon entropy | 0.42546 |
| G+C content | 0.42123 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -11.92 |
| Energy contribution | -13.07 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
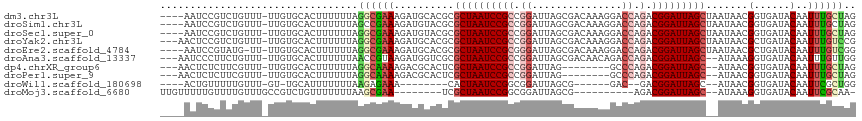
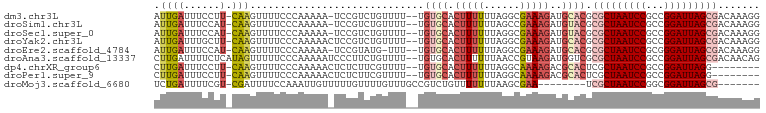
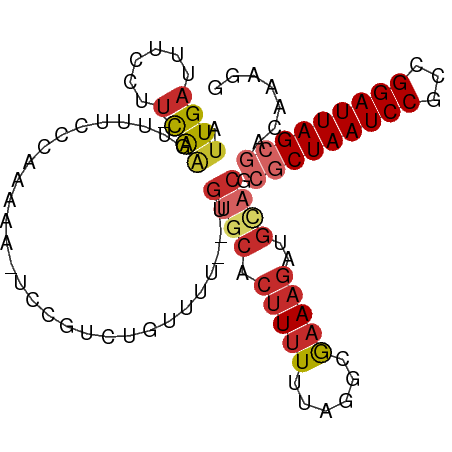
>dm3.chr3L 14166404 97 - 24543557 AUUGAUUUCCUU-CAAGUUUUCCCAAAAA-UCCGUCUGUUUU--UGUGCACUUUUUUAGGCGAAAGAUGCACGCGCUAAUCCGCCGGAUUAGCGACAAAGG .((((......)-))).............-...(((......--.((((((((((......))))).)))))..((((((((...)))))))))))..... ( -25.80, z-score = -1.93, R) >droSim1.chr3L 13522484 97 - 22553184 AUUGAUUUCCAU-CAAGUUUUCCCAAAAA-UCCGUCUGUUUU--UGUGCACUUUUUUAGCCGAAAGAUGUACGCGCUAAUCCGCCGGAUUAGCGACAAAGG .(((((....))-))).............-...(((......--.((((((((((......))))).)))))..((((((((...)))))))))))..... ( -23.70, z-score = -2.15, R) >droSec1.super_0 6313459 97 - 21120651 AUUGAUUUCCAU-CAAGUUUUCCCAAAAA-UCCGUCUGUUUU--UGUGCACUUUUUUAGGCGAAAGAUGUACGCGCUAAUCCGCCGGAUUAGCGACAAAGG .(((((....))-))).............-...(((......--.((((((((((......))))).)))))..((((((((...)))))))))))..... ( -23.70, z-score = -1.57, R) >droYak2.chr3L 14260971 98 - 24197627 AUUGAUUUGCUU-CAAGUUUUCCCAAAAACUCCGUCUGUUUU--UGUGCACUUUUUUAGGCGAAAGAUGCACGCGCUAAUCCGCCGGAUUAGCGACAAAGG .((((......)-)))......((((((((.......)))))--)((((((((((......))))).)))))((((((((((...))))))))).)...)) ( -27.20, z-score = -1.90, R) >droEre2.scaffold_4784 14157188 96 - 25762168 AUUGAUUUCCAU-CAAGUUUUCCCAAAAA-UCCGUAUG-UUU--UGUGCACUUUUUUAGGCGAAAGAUGCACGCGCUAAUCCGCGGGAUUAGCGACAAAGG .(((((....))-))).............-.((...((-(..--.((((((((((......))))).))))).(((((((((...))))))))))))..)) ( -25.10, z-score = -1.51, R) >droAna3.scaffold_13337 18411670 99 + 23293914 CUUGAUUUUCUCAUAGUUUUUCCCAAAAAUCCCUUCUGUUUU--UGUGCACUUUUUUAACCGUAAGAUGGUCGCGCUAAUCCGCCGGAUUAGCGACAACAG ...((((((................))))))....(((((.(--((((.(((.(((((....))))).))))))((((((((...)))))))))).))))) ( -19.49, z-score = -1.50, R) >dp4.chrXR_group6 4854373 90 - 13314419 CUUGAUUUCCUU-CAAGUUUUCCCAAAAACUCUCUUCGUUUU--UGUGCACUUUUUUAGGCAAAAGACGCACUCGCUAAUCCGCCGGAUUAGG-------- (((((......)-))))...................((((((--(.(((.((.....))))))))))))......(((((((...))))))).-------- ( -18.90, z-score = -1.65, R) >droPer1.super_9 3156506 90 - 3637205 CUUGAUUUCCUU-CAAGUUUUCCCAAAAACUCUCUUCGUUUU--UGUGCACUUUUUUAGGCAAAAGACGCACUCGCUAAUCCGCCGGAUUAGG-------- (((((......)-))))...................((((((--(.(((.((.....))))))))))))......(((((((...))))))).-------- ( -18.90, z-score = -1.65, R) >droMoj3.scaffold_6680 19236969 85 - 24764193 UCUGAUUUUCGU-CGAUUUUCCAAAUUGUUUUUGUUUUGUUUGCCGUCUGUUUUUUUAAGCGAA--------UCGCUAAUCCGGCGGAUUAGCG------- ...(((((..((-.(((....((((.....))))....))).)).....((((....)))))))--------))((((((((...)))))))).------- ( -16.10, z-score = -0.37, R) >consensus AUUGAUUUCCUU_CAAGUUUUCCCAAAAA_UCCGUCUGUUUU__UGUGCACUUUUUUAGGCGAAAGAUGCACGCGCUAAUCCGCCGGAUUAGCGACAAAGG .............................................((((..(((((.....)))))..)))).(((((((((...)))))))))....... (-11.92 = -13.07 + 1.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:11 2011