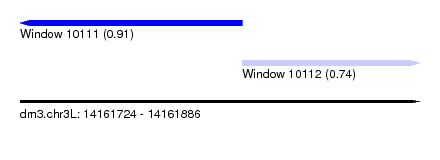
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,161,724 – 14,161,886 |
| Length | 162 |
| Max. P | 0.908562 |

| Location | 14,161,724 – 14,161,814 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 66.78 |
| Shannon entropy | 0.64269 |
| G+C content | 0.43973 |
| Mean single sequence MFE | -16.77 |
| Consensus MFE | -7.61 |
| Energy contribution | -8.24 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
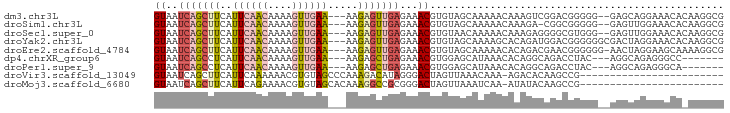
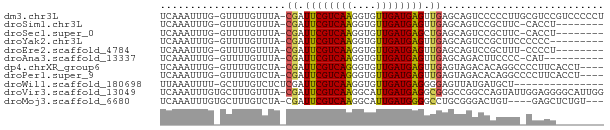
>dm3.chr3L 14161724 90 - 24543557 GUAAUCAGCUUCAUUCAACAAAAGUUGAA---AAGAGUUGAGAAACGUGUAGCAAAAACAAAGUCGGACGGGGG--GAGCAGGAAACACAAGGCG ....(((((((..((((((....))))))---..)))))))....(((.(..(.........)..).)))....--..((.(....).....)). ( -18.80, z-score = -1.99, R) >droSim1.chr3L 13517764 89 - 22553184 GUAAUCAGCUUCAUUCAACAAAAGUUGAA---AAGAGUUGAGAAACGUGUAGCAAAAACAAAGA-CGGCGGGGG--GAGUUGGAAACACAAGGCG ((..(((((((..((((((....))))))---..)))))))...))((((.(......).....-((((.....--..))))...))))...... ( -18.90, z-score = -1.95, R) >droSec1.super_0 6308735 90 - 21120651 GUAAUCAGCUUCAUUCAACAAAAGUUGAA---AAGAGUUGAGAAACGUGUAACAAAAACAAAGAGGGGCGUGGG--GAGUUGGAAACACAAGGCG ((..(((((((..((((((....))))))---..)))))))...))((((..(((...(...(.....)....)--...)))...))))...... ( -18.70, z-score = -1.92, R) >droYak2.chr3L 14255790 92 - 24197627 GUAAUCAGCUUCAUUCAACAAAAGUUGAA---AAGAGUUGAGAAACGUGUAGCAAAAGCACAGAUGGACGGGGGGCGACUAGGAAACACAAGGCG ((..(((((((..((((((....))))))---..)))))))...))((((.((....)).....(((.((.....)).)))....))))...... ( -21.10, z-score = -2.20, R) >droEre2.scaffold_4784 14152284 91 - 25762168 GUAAUCAGCUUCAUUCAACAAAAGUUGAA---AAGAGUUGAGAAACGUGUAGCAAAAACACAGACGAACGGGGGG-AACUAGGAAGCAAAAGGCG ((..(((((((..((((((....))))))---..)))))))...))((((.......)))).......(..((..-..))..)..((.....)). ( -19.90, z-score = -2.57, R) >dp4.chrXR_group6 4849784 82 - 13314419 GUAAUCAGCCUCAUUCAACAAAAGUUGAA---AAGAGCUGAGAAACGUGGAGCAUAAACACAGGCAGACCUAC---AGGCAGAGGGCC------- ((..(((((((..((((((....))))))---.)).)))))...))(((.........)))(((....)))..---.(((.....)))------- ( -20.10, z-score = -1.79, R) >droPer1.super_9 3151903 82 - 3637205 GUAAUCAGCCUCAUUCAACAAAAGUUGAA---AAGAGCUGAGAAACGUGGAGCAUAAACACAGGCAGACCUAC---AGGCAGAGGGCA------- ........((((.((((((....))))))---....(((.......(((.........)))(((....)))..---.))).))))...------- ( -18.60, z-score = -1.26, R) >droVir3.scaffold_13049 18434872 70 - 25233164 GUAAUCAGCUUCAUUCAAAAAACGUGUAGCCCAAAGACAUAGGGACUAGUUAAACAAA-AGACACAAGCCG------------------------ .......((((...............((((((.........))).)))(((.......-.)))..))))..------------------------ ( -8.20, z-score = -0.38, R) >droMoj3.scaffold_6680 19232532 70 - 24764193 GUAAUCAGCUUCAUUCAGAAAACGUGUAGCACAAAGGCCGCGGGACUAGUUAAAUCAA-AUAUACAAGCCG------------------------ ......................((((..((......))))))((.((.((........-....)).)))).------------------------ ( -6.60, z-score = 1.41, R) >consensus GUAAUCAGCUUCAUUCAACAAAAGUUGAA___AAGAGUUGAGAAACGUGUAGCAAAAACACAGACAGACGGGGG__GAGUAGGAAACA_AAGGCG ((..(((((((..((((((....)))))).....)))))))...))................................................. ( -7.61 = -8.24 + 0.63)
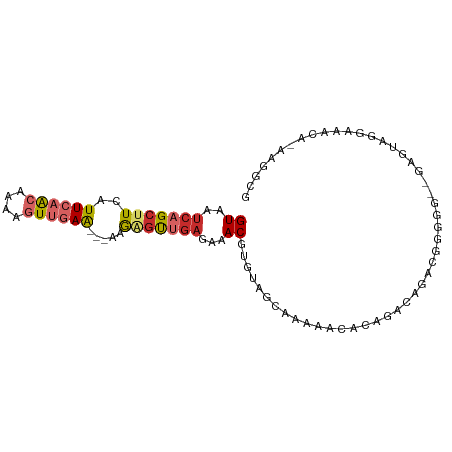
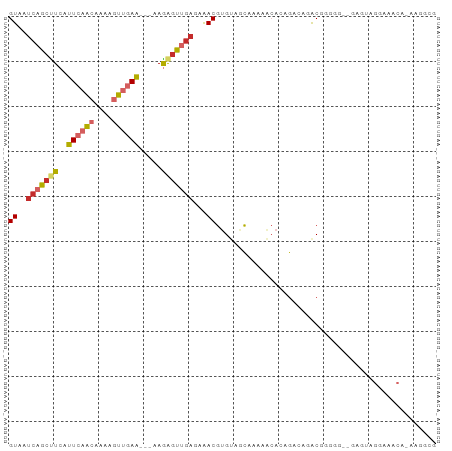
| Location | 14,161,814 – 14,161,886 |
|---|---|
| Length | 72 |
| Sequences | 11 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 71.87 |
| Shannon entropy | 0.60579 |
| G+C content | 0.45209 |
| Mean single sequence MFE | -17.21 |
| Consensus MFE | -6.64 |
| Energy contribution | -6.46 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.741018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
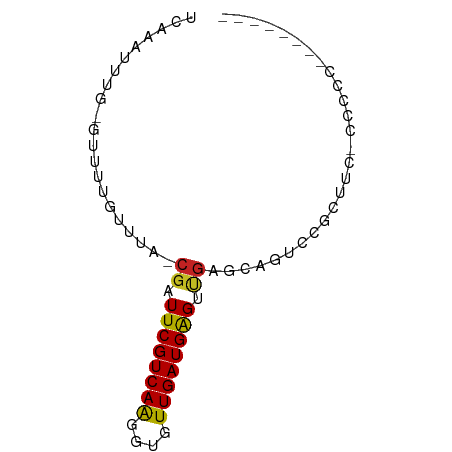
>dm3.chr3L 14161814 72 + 24543557 UCAAAUUUG-GUUUUGUUUA-CGAUUCGUCAAGGUGUUGAUGAGUUGAGCAGUCCCCCUUGCGUCCGUCCCCCU ........(-(..(((((..-(((((((((((....))))))))))))))))..)).................. ( -16.70, z-score = -2.06, R) >droSim1.chr3L 13517853 63 + 22553184 UCAAAUUUG-GUUUUGUUUA-CGAUUCGUCAAGGUGUUGAUGAGUUGAGCAGUCCGCUUC-CACCU-------- ........(-(..(((((..-(((((((((((....)))))))))))))))).)).....-.....-------- ( -15.40, z-score = -1.41, R) >droSec1.super_0 6308825 63 + 21120651 UCAAAUUUG-GUUUUGUUUA-CGAUUCGUCAAGGUGUUGAUGAGCUGAGCAGUCCGCUUC-CACCU-------- ........(-(..(((((((-.(.((((((((....)))))))))))))))).)).....-.....-------- ( -13.90, z-score = -0.52, R) >droYak2.chr3L 14255882 63 + 24197627 UCAAAUUUG-GUUUUGUUUA-CGAUUCGUCAAGGUGUUGAUGAGUUGAGCAGUCCGCUUCCCCCC--------- ........(-(..(((((..-(((((((((((....)))))))))))))))).))..........--------- ( -15.40, z-score = -2.03, R) >droEre2.scaffold_4784 14152375 63 + 25762168 UCAAAUUUG-GUUUUGUUUA-CGAUUCGUCAAGGUGUUGAUGAGUUGAGCAGUCCGCUUU-CCCCU-------- ........(-(..(((((..-(((((((((((....)))))))))))))))).)).....-.....-------- ( -15.40, z-score = -1.81, R) >droAna3.scaffold_13337 18407676 61 - 23293914 UCAAAUUUG-GUUUUGUUUA-CGAUUCGUCAAGGUGUUGAUGAGUUGAGCAGACUUCCCC-CAU---------- ........(-(.((((((..-(((((((((((....)))))))))))))))))...))..-...---------- ( -15.10, z-score = -1.75, R) >dp4.chrXR_group6 4849866 68 + 13314419 UCAAAUUUG-GUUUUGUCUA-CGAUUCGUCAGGGUGUUGAUGAGUUGAGUAGACACAGGCCCCUUCACCU---- ........(-((((((((((-(((((((((((....))))))))))..))))))).))))).........---- ( -22.10, z-score = -2.29, R) >droPer1.super_9 3151985 68 + 3637205 UCAAAUUUG-GUUUUGUCUA-CGAUUCGUCAGGGUGUUGAUGAGUUGAGUAGACACAGGCCCCUUCACCU---- ........(-((((((((((-(((((((((((....))))))))))..))))))).))))).........---- ( -22.10, z-score = -2.29, R) >droWil1.scaffold_180698 1361591 58 + 11422946 UUAAAUUUU-GCUUUGUCUCUCGAUUCGUCAAGGUGUUGAUGAGGGGAGUUAUGAUGCU--------------- .........-((.(..(..(((..((((((((....))))))))..)))..)..).)).--------------- ( -15.00, z-score = -2.01, R) >droVir3.scaffold_13049 18434942 73 + 25233164 UCAAAUUUGUGCUUUGUUUA-CGAUUCGUCAAGGCAUUGAUGAGGCGGGCCGGCCAGUAUUGGAGGGGCAUUGG .......(((.((((...((-(...(((((((....)))))))(((......))).)))...)))).))).... ( -19.40, z-score = -0.34, R) >droMoj3.scaffold_6680 19232602 66 + 24764193 UCAAAUUUGUGCUUUGUCUA-CGAUUCGUCAAGGCAUUGAUGGGGCCUGCGGGACUGU----GAGCUCUGU--- ....(((.((((((((.(..-......).)))))))).)))(((((..(((....)))----..)))))..--- ( -18.80, z-score = -0.80, R) >consensus UCAAAUUUG_GUUUUGUUUA_CGAUUCGUCAAGGUGUUGAUGAGUUGAGCAGUCCGCUUC_CCCCC________ .....................((.((((((((....)))))))).))........................... ( -6.64 = -6.46 + -0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:08 2011