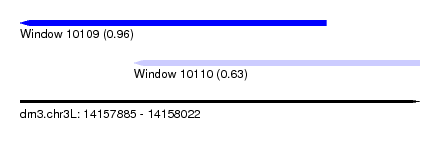
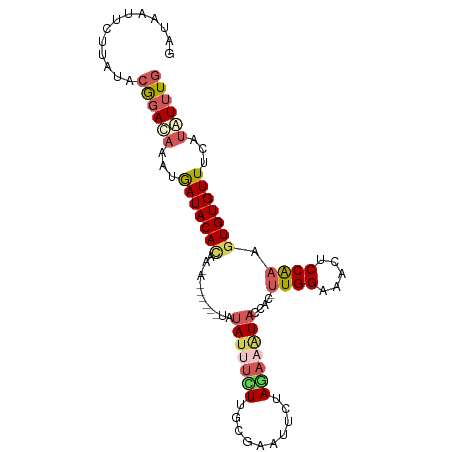
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,157,885 – 14,158,022 |
| Length | 137 |
| Max. P | 0.962710 |

| Location | 14,157,885 – 14,157,990 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 71.10 |
| Shannon entropy | 0.52546 |
| G+C content | 0.34173 |
| Mean single sequence MFE | -20.76 |
| Consensus MFE | -11.48 |
| Energy contribution | -12.76 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
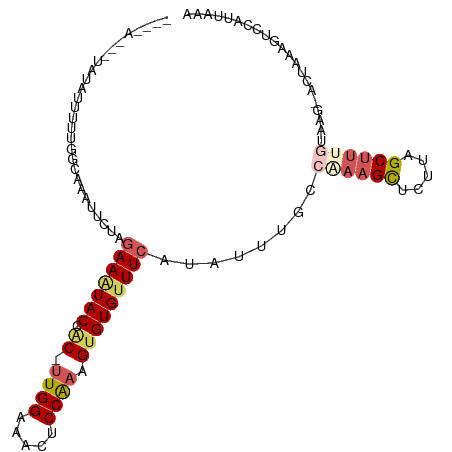
>dm3.chr3L 14157885 105 - 24543557 --AAAGAUUAUAUUUUUGAUGAAUUCUAGAAAUACCAC-UUGGAAACUCCAAAGUGUGUUUCAUAUUUGCCAAAGCUCUUAGCUUUGUAGGUCCUAAGGUCUAUUAAA --............(((((((.((..(((((((((.((-((((.....)).)))))))))))..((((((.(((((.....)))))))))))..))..)).))))))) ( -22.70, z-score = -1.70, R) >droSim1.chr3L 13513860 98 - 22553184 --------UAUAUUUCUUGCAAAUUCUAGAAAUACCAC-UUGGAAACUCCAAAGUGUGUUUCAUAUUUGCCGAAGCUCUUAGCUAAGUAAG-ACUAAAGUCCAUUAAA --------......((((((........(((((((.((-((((.....)).)))))))))))...........(((.....)))..)))))-)............... ( -20.80, z-score = -2.60, R) >droSec1.super_0 6304862 98 - 21120651 --------UAUAUUUCUUGCACAUUCUAGAAAUACCAC-CUGGAAACUCCAAAGUGUGUUUCAUAUUUGCCGAAGCUCUUAGCUUUGGAAG-ACUAAAGUCCAUUAAA --------....................(((((((.((-.(((.....)))..))))))))).......(((((((.....)))))))..(-((....)))....... ( -21.50, z-score = -2.19, R) >droYak2.chr3L 14251183 107 - 24197627 AACGAUUGUGCAUACUUGUAGUAUUAUAAAAGUACCUCUUUGGAAAUUCCGAAGUGUGUUUCACGUUUGCCCAAGUUCUUAGCUUCAUAAG-GCUGAGUGCCAUUAAA .....(((.(((.((.((..(((((.....)))))(.((((((.....)))))).).....)).)).))).)))((.(((((((......)-)))))).))....... ( -24.10, z-score = -1.51, R) >droEre2.scaffold_4784 14148411 95 - 25762168 AACUAUUAUACAUAUUU------UGAAAGAAUUACCGCUUUGGAAAUUCCGAAGUGUGUUUCAUGUUUACCAAAGCUCUUAGCCUUACGAA-GC------CCAUUAAA ..............(((------((.(((......((((((((.....)))))))).(((....((((....))))....)))))).))))-).------........ ( -14.70, z-score = -0.94, R) >consensus ____A___UAUAUUUUUGGCAAAUUCUAGAAAUACCAC_UUGGAAACUCCAAAGUGUGUUUCAUAUUUGCCAAAGCUCUUAGCUUUGUAAG_ACUAAAGUCCAUUAAA ............................(((((((.((.((((.....)))).)))))))))........((((((.....))))))..................... (-11.48 = -12.76 + 1.28)

| Location | 14,157,924 – 14,158,022 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 75.15 |
| Shannon entropy | 0.44378 |
| G+C content | 0.29634 |
| Mean single sequence MFE | -16.54 |
| Consensus MFE | -9.32 |
| Energy contribution | -9.64 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
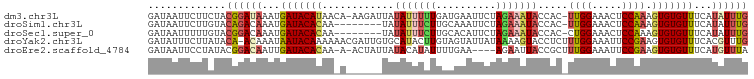
>dm3.chr3L 14157924 98 - 24543557 GAUAAUUCUUCUACGGAUAAAUGAUACAUAACA-AAGAUUAUAUUUUUGAUGAAUUCUAGAAAUACCAC-UUGGAAACUCCAAAGUGUGUUUCAUAUUUG ...(((((.((....))..............((-(((((...)))))))..)))))...(((((((.((-((((.....)).)))))))))))....... ( -17.70, z-score = -1.64, R) >droSim1.chr3L 13513898 91 - 22553184 GAUAAUUCUUGUACAGACAAAUGAUACACAA--------UAUAUUUCUUGCAAAUUCUAGAAAUACCAC-UUGGAAACUCCAAAGUGUGUUUCAUAUUUG ........((((....))))((((.(((((.--------..(((((((..........)))))))....-((((.....))))..))))).))))..... ( -16.20, z-score = -1.74, R) >droSec1.super_0 6304900 91 - 21120651 GAUAAUUUUUGUACGGACAAAUGAUACACAA--------UAUAUUUCUUGCACAUUCUAGAAAUACCAC-CUGGAAACUCCAAAGUGUGUUUCAUAUUUG ........((((....))))((((.(((((.--------..(((((((..........)))))))....-.(((.....)))...))))).))))..... ( -15.40, z-score = -1.23, R) >droYak2.chr3L 14251221 99 - 24197627 GAUAUUUCUUAUACA-ACAAAUAAUACAAAAAACGAUUGUGCAUACUUGUAGUAUUAUAAAAGUACCUCUUUGGAAAUUCCGAAGUGUGUUUCACGUUUG ...............-..............(((((.....(((((((((..(((((.....)))))..)..(((.....)))))))))))....))))). ( -15.70, z-score = -0.70, R) >droEre2.scaffold_4784 14148443 94 - 25762168 GAUAAUUCCUAUACGGACAAUUGAUACACAA-A-ACUAUUAUACAUAUUUUGAA----AGAAUUACCGCUUUGGAAAUUCCGAAGUGUGUUUCAUGUUUA ..............(((((..(((.((((..-.-...........((.(((...----.))).))..(((((((.....))))))))))).)))))))). ( -17.70, z-score = -1.87, R) >consensus GAUAAUUCUUAUACGGACAAAUGAUACACAA_A______UAUAUUUCUUGCGAAUUCUAGAAAUACCAC_UUGGAAACUCCAAAGUGUGUUUCAUAUUUG ...........................................................(((((((.((.((((.....)))).)))))))))....... ( -9.32 = -9.64 + 0.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:07 2011