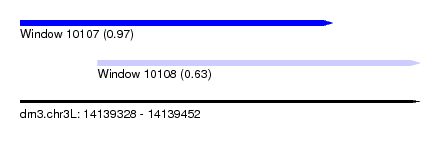
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,139,328 – 14,139,452 |
| Length | 124 |
| Max. P | 0.974527 |

| Location | 14,139,328 – 14,139,425 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.64 |
| Shannon entropy | 0.50057 |
| G+C content | 0.39486 |
| Mean single sequence MFE | -27.09 |
| Consensus MFE | -15.06 |
| Energy contribution | -14.70 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
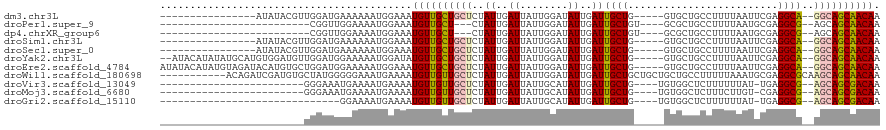
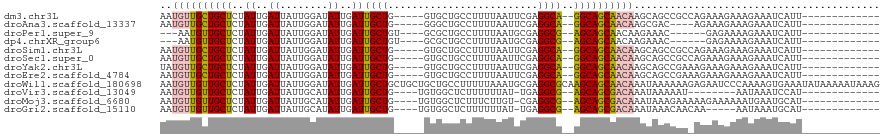
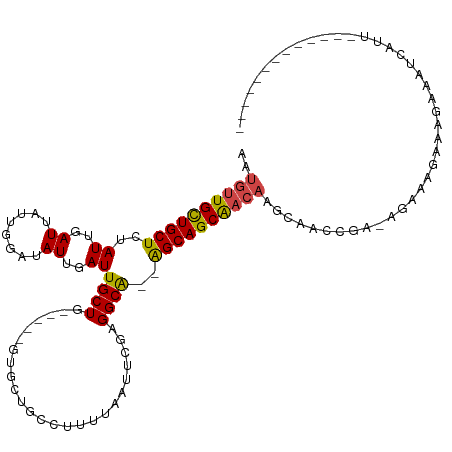
>dm3.chr3L 14139328 97 + 24543557 ----------------AUAUACGUUGGAUGAAAAAAUGGAAAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG-----GUGCUGCCUUUUAAUUCGAGGCA--GGCAGCAACAA ----------------.....((((.........))))....(((((((((.(((..(((((........)))))..))-----)..(((((((.......))))))--)))))))))). ( -28.10, z-score = -2.67, R) >droPer1.super_9 3128128 86 + 3637205 -------------------------CGGUUGGAAAAUGGAAAUGUUGCU---CUAUUGAUUAUUGGAUAUUGAUUGCUGU----GCGCUGCCUUUUAAUGCGAGGCG--AGCAGCAACAA -------------------------(.(((....))).)...(((((((---(....(((((........)))))(((..----.(((...........))).))))--))))))).... ( -22.20, z-score = -0.72, R) >dp4.chrXR_group6 4826290 86 + 13314419 -------------------------CGGUUGGAAAAUGGAAAUGUUGCU---CUAUUGAUUAUUGGAUAUUGAUUGCUGU----GCGCUGCCUUUUAAUGCGAGGCG--AGCAGCAACAA -------------------------(.(((....))).)...(((((((---(....(((((........)))))(((..----.(((...........))).))))--))))))).... ( -22.20, z-score = -0.72, R) >droSim1.chr3L 13494279 97 + 22553184 ----------------AUAUACGUUGGAUGAAAAAAUGGAAAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG-----GUGCUGCCUUUUAAUUCGAGGCA--GGCAGCAACAA ----------------.....((((.........))))....(((((((((.(((..(((((........)))))..))-----)..(((((((.......))))))--)))))))))). ( -28.10, z-score = -2.67, R) >droSec1.super_0 6286848 97 + 21120651 ----------------AUAUACGUUGGAUGAAAAAAUGGAAAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG-----GUGCUGCCUUUUAAUUCGAGGCA--GGCAGCAACAA ----------------.....((((.........))))....(((((((((.(((..(((((........)))))..))-----)..(((((((.......))))))--)))))))))). ( -28.10, z-score = -2.67, R) >droYak2.chr3L 14231673 111 + 24197627 --AUACAUAUAUGCAUGUGGAUGUUGGAUGGAAAAAUGGAUAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG-----GUGCUGCCUUUUAAUUCGAGGCA--GGCAGCAACAA --...(((....((((....))))...)))............(((((((((.(((..(((((........)))))..))-----)..(((((((.......))))))--)))))))))). ( -31.10, z-score = -1.62, R) >droEre2.scaffold_4784 14130129 113 + 25762168 AUAUACAUAUGUAGAUACAUGUGCUGGAUGGAAAAAUGGAAAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG-----GUGCUGCCUUUUAAUUCGAGGCA--GGCAGCAACAA .....(((((((....)))))))...................(((((((((.(((..(((((........)))))..))-----)..(((((((.......))))))--)))))))))). ( -33.30, z-score = -2.49, R) >droWil1.scaffold_180698 10036378 109 - 11422946 -----------ACAGAUCGAUGUGCUAUGGGGGAAAUGAAAAUGUUGUUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUGCUGCUGCUGCCUUUUUAAAUGCGAGGCGCAAGCAGCAACAA -----------...(((((((((.(.((((((..((..(.....)..)).))))))........).)))))))))(.(((((((((.(((((.........))))))).))))))).).. ( -32.00, z-score = -1.46, R) >droVir3.scaffold_13049 18413810 89 + 25233164 ------------------------GGGAAAUGAAAAUGAAAAUGUUGUUGCUCUAUUGAUUAUUGCAUAUUGAUUGCUG----UGUGGCUCUUUUUUAU-UGAGGCG--AGCAGCGACAA ------------------------..................(((((((((((.......(((.(((.......))).)----)).(.(((........-.))).))--)))))))))). ( -23.80, z-score = -2.21, R) >droMoj3.scaffold_6680 19207697 89 + 24764193 ------------------------GGGAAAUGAAAAUGAAAAUGUUGUUGCUCUAUUGAUUAUUGCAUAUUGAUUGCUG----UGUGGCUCUUUCUUGU-CGAGGCG--AGCAGCGACAA ------------------------..................(((((((((((....(((((........)))))(((.----..((((........))-)).))))--)))))))))). ( -25.30, z-score = -2.11, R) >droGri2.scaffold_15110 15331253 83 - 24565398 ------------------------------GGAAAAUGAAAAUGUUGUUGCUCUAUUGAUUAUUGCAUAUUGAUUGCUG----UGUGGCUCUUUUUUAU-UGAGGCG--AGCAGCGACAA ------------------------------............(((((((((((.......(((.(((.......))).)----)).(.(((........-.))).))--)))))))))). ( -23.80, z-score = -2.56, R) >consensus ________________________UGGAUGGGAAAAUGGAAAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG_____GUGCUGCCUUUUAAUUCGAGGCG__AGCAGCAACAA ..........................................((((((((((.....(((((........)))))...............((((.......))))....)))))))))). (-15.06 = -14.70 + -0.36)
| Location | 14,139,352 – 14,139,452 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.51 |
| Shannon entropy | 0.44573 |
| G+C content | 0.38445 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -15.30 |
| Energy contribution | -14.82 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626467 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
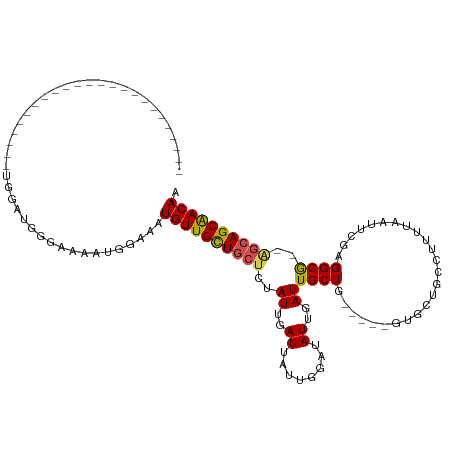
>dm3.chr3L 14139352 100 + 24543557 AAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG-----GUGCUGCCUUUUAAUUCGAGGCA--GGCAGCAACAAGCAGCCGCCAGAAAGAAAGAAAUCAUU------------- ...((.(((((((((........))))..(((.((((((-----...(((((((.......))))))--).)))))))))))))).))...................------------- ( -29.60, z-score = -1.17, R) >droAna3.scaffold_13337 18386926 96 - 23293914 AAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG-----GGGCUGCCUUUUAAUUCGAGGCA--GGCAGCAACAAGCGAC----AGAAAGAAAGAAAUCAUU------------- ..(((((((((((((..(((((........)))))..))-----)).(((((((.......))))))--))))))))))......----..................------------- ( -28.80, z-score = -2.08, R) >droPer1.super_9 3128143 92 + 3637205 ---AAUGUUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUGU----GCGCUGCCUUUUAAUGCGAGGCG--AGCAGCAACAAGAAAC------GAGAAAAGAAAUCAUU------------- ---..((((((((....(((((........)))))(((..----.(((...........))).))))--))))))).........------................------------- ( -20.50, z-score = -0.14, R) >dp4.chrXR_group6 4826305 92 + 13314419 ---AAUGUUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUGU----GCGCUGCCUUUUAAUGCGAGGCG--AGCAGCAACAAGAAAC------GAGAAAAGAAAUCAUU------------- ---..((((((((....(((((........)))))(((..----.(((...........))).))))--))))))).........------................------------- ( -20.50, z-score = -0.14, R) >droSim1.chr3L 13494303 100 + 22553184 AAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG-----GUGCUGCCUUUUAAUUCGAGGCA--GGCAGCAACAAGCAGCCGCCAGAAAGAAAGAAAUCAUU------------- ...((.(((((((((........))))..(((.((((((-----...(((((((.......))))))--).)))))))))))))).))...................------------- ( -29.60, z-score = -1.17, R) >droSec1.super_0 6286872 100 + 21120651 AAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG-----GUGCUGCCUUUUAAUUCGAGGCA--GGCAGCAACAAGCAGCCGCCAGAAAGAAAGAAAUCAUU------------- ...((.(((((((((........))))..(((.((((((-----...(((((((.......))))))--).)))))))))))))).))...................------------- ( -29.60, z-score = -1.17, R) >droYak2.chr3L 14231711 100 + 24197627 UAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG-----GUGCUGCCUUUUAAUUCGAGGCA--GGCAGCAACAAGCAGCCGAAAGAAAGAAAGAAAUCAUU------------- ......(((((((((........))))..(((.((((((-----...(((((((.......))))))--).))))))))))))))(....)................------------- ( -29.20, z-score = -1.67, R) >droEre2.scaffold_4784 14130169 100 + 25762168 AAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG-----GUGCUGCCUUUUAAUUCGAGGCA--GGCAGCAACAAGCAGCCGAAAGAAAGAAAGAAAUCAUU------------- ......(((((((((........))))..(((.((((((-----...(((((((.......))))))--).))))))))))))))(....)................------------- ( -29.20, z-score = -1.60, R) >droWil1.scaffold_180698 10036407 120 - 11422946 AAUGUUGUUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUGCUGCUGCUGCCUUUUUAAAUGCGAGGCGCAAGCAGCAACAAAUAAAAAAGAGAAUCCCAAAAGUGAAAUAUAAAAAUAAAG .(((((.(..((.....((((.((...((((..((((((((..(((.(((((.........))))))))))))))))..))))...))...)))).....))..)))))).......... ( -27.90, z-score = -1.47, R) >droVir3.scaffold_13049 18413826 92 + 25233164 AAUGUUGUUGCUCUAUUGAUUAUUGCAUAUUGAUUGCUG----UGUGGCUCUUUUUUAU-UGAGGCG--AGCAGCGACAAAUAAAAAU--------AAUAAAUCCAU------------- ..(((((((((((.......(((.(((.......))).)----)).(.(((........-.))).))--)))))))))).........--------...........------------- ( -23.80, z-score = -2.23, R) >droMoj3.scaffold_6680 19207713 100 + 24764193 AAUGUUGUUGCUCUAUUGAUUAUUGCAUAUUGAUUGCUG----UGUGGCUCUUUCUUGU-CGAGGCG--AGCAGCGACAAAUAAAGAAAAAGAAAAAAUGAAUGCAU------------- ..(((((((((((....(((((........)))))(((.----..((((........))-)).))))--))))))))))............................------------- ( -25.30, z-score = -1.55, R) >droGri2.scaffold_15110 15331263 95 - 24565398 AAUGUUGUUGCUCUAUUGAUUAUUGCAUAUUGAUUGCUG----UGUGGCUCUUUUUUAU-UGAGGCG--AGCAGCGACAAAUAAACAACAA-----AAUAAAUGCAU------------- ..(((((((((..((.....))..)).((((..((((((----(.((.(((........-.))).))--.)))))))..))))))))))).-----...........------------- ( -25.00, z-score = -1.85, R) >consensus AAUGUUGCUGCUCUAUUGAUUAUUGGAUAUUGAUUGCUG_____GUGCUGCCUUUUAAUUCGAGGCA__AGCAGCAACAAGCAACCGA_AGAAAGAAAGAAAUCAUU_____________ ..((((((((((.....(((((........)))))...............((((.......))))....))))))))))......................................... (-15.30 = -14.82 + -0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:05 2011