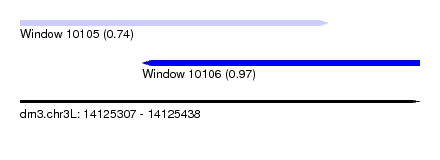
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,125,307 – 14,125,438 |
| Length | 131 |
| Max. P | 0.966746 |

| Location | 14,125,307 – 14,125,408 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.66 |
| Shannon entropy | 0.46799 |
| G+C content | 0.49597 |
| Mean single sequence MFE | -30.95 |
| Consensus MFE | -19.57 |
| Energy contribution | -19.60 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.744860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14125307 101 + 24543557 GCAACUACGAUUCGCCUUUGGCCAAUUCCCAAGCUCAAAAUGGUGGAUAUGUUAACCAAGGUGGCCAUA--GUCCAAAAUUGGUCCAGACAGACCACUUUGGG----------------- ..(((....(((((((((((((..........)).))))..)))))))..)))..((((((((((....--(.(((....))).)......).))))))))).----------------- ( -26.10, z-score = -0.41, R) >droAna3.scaffold_13337 18373946 116 - 23293914 ----CUGCGAAUCGCCUUCGGCUAAUUUCUAAGCUCAAAAUGGCGGAUAUGUUGGCCAAGGUGGGAAUGUGAAGCCAAAUGGCCGCCAAUGGAGGUCCAAAGAGGUCGAUGGAUGGUAGA ----((((.(....((.((((((..(((...(.(((....((((((.....(((((..(.((....)).)...)))))....))))))...))).)..)))..)))))).)).).)))). ( -36.20, z-score = -0.65, R) >droEre2.scaffold_4784 14115466 90 + 25762168 GCAACUACGAUUCGCCUUUGGCUAAUUUCCAAGCUCAAAAUGGUGGAUAUGUUGGCCAAGGUGGCCAUA--AUACGAG-----------CAGAACGCUUUGGA----------------- .((((....((((((((((((((........))).))))..)))))))..)))).((((((((..(...--.......-----------..)..)))))))).----------------- ( -26.20, z-score = -1.18, R) >droYak2.chr3L 14216838 91 + 24197627 GCAACUGCGAUUCGCCUUUGGCCAAUUCGCAAGCUCAAAAUGGUGGAUAUGUUGGCCAAGGUGGCCAUA--AUCCGAAAUGGGUCUCGACAGA--------------------------- ....((((((.(((((((.(((((((......(((......)))......))))))))))))))((((.--.......))))...))).))).--------------------------- ( -30.20, z-score = -1.33, R) >droSec1.super_0 6272653 101 + 21120651 GCAACUACGAUUCGCCUUUGGCCAAUUCCCAAGCUCAAAAUGGUGGAUAUGUUGGCCAAGGUGGCCAUA--GUCCGAAAUGCGUCUCGAUAGACCAUUUUGGG----------------- ...((((.(..(((((((.((((((((((...(((......))))))...)))))))))))))).).))--))((((((((.((((....)))))))))))).----------------- ( -35.80, z-score = -2.67, R) >droSim1.chr3L 13480026 101 + 22553184 GCAACUACGAUUCGCCUUUGGCCAAUUCCCAAGCUCAAAAUGGUGGAUAUGUUGGCCAAGGUGGCCAUA--GUCCGAAAUGCGUCCCGACAGACCACUUUGGG----------------- ...((((.(..(((((((.((((((((((...(((......))))))...)))))))))))))).).))--))(((((.((.(((......))))).))))).----------------- ( -31.20, z-score = -0.99, R) >consensus GCAACUACGAUUCGCCUUUGGCCAAUUCCCAAGCUCAAAAUGGUGGAUAUGUUGGCCAAGGUGGCCAUA__AUCCGAAAUGGGUCCCGACAGACCACUUUGGG_________________ ...........(((((((.(((((((......(((......)))......))))))))))))))........................................................ (-19.57 = -19.60 + 0.03)

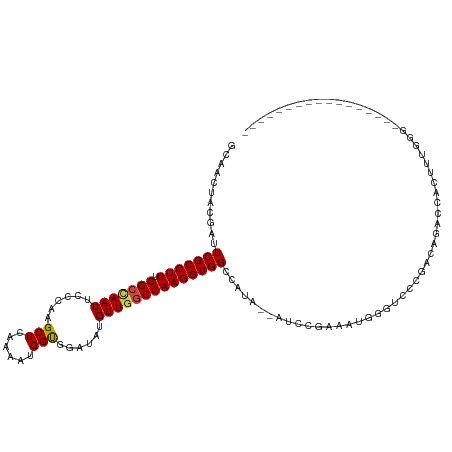
| Location | 14,125,347 – 14,125,438 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 70.60 |
| Shannon entropy | 0.52765 |
| G+C content | 0.44581 |
| Mean single sequence MFE | -23.44 |
| Consensus MFE | -11.56 |
| Energy contribution | -11.68 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14125347 91 - 24543557 --UAAUCUUUGUCUGAGCCAAAGAUAAAGUGUCCCAAAGUGGUCUGUCUGGACCAAUUUUGGACUAUGGCCACCUUGGUUAACAUAUCCACCA --....((((((((.......))))))))(((.((((.((((((.(((..((.....))..)))...)))))).))))...)))......... ( -27.70, z-score = -2.54, R) >droEre2.scaffold_4784 14115506 89 - 25762168 UAUAACUGUGUUAGAACCUACAACCGAAAU---AUAAAGUGUUCCAAAGCGUUCUG-CUCGUAUUAUGGCCACCUUGGCCAACAUAUCCACCA ......(((((((((((((..((((.....---.....).)))....)).))))))-.........(((((.....))))))))))....... ( -15.40, z-score = -0.68, R) >droYak2.chr3L 14216878 71 - 24197627 ------------------UAAGUCUAAAA----ACAAAGUGGUCUGUCGAGACCCAUUUCGGAUUAUGGCCACCUUGGCCAACAUAUCCACCA ------------------...........----.....(.(((((....)))))).....((((..(((((.....)))))....)))).... ( -19.90, z-score = -2.56, R) >droSec1.super_0 6272693 91 - 21120651 --UAAUCUUUGUGUGCGCUAAAGAUAAAGUGUCCCAAAAUGGUCUAUCGAGACGCAUUUCGGACUAUGGCCACCUUGGCCAACAUAUCCACCA --..((((((((....)).)))))).....((((..(((((((((....)))).))))).))))..(((((.....)))))............ ( -25.50, z-score = -2.45, R) >droSim1.chr3L 13480066 91 - 22553184 --UAAUCUUUGUGUGCGCUAAAGAUAAAGUGUCCCAAAGUGGUCUGUCGGGACGCAUUUCGGACUAUGGCCACCUUGGCCAACAUAUCCACCA --..((((((((....)).))))))...(((((((..((....))...))))))).....(((...(((((.....))))).....))).... ( -28.70, z-score = -2.06, R) >consensus __UAAUCUUUGUGUGAGCUAAAGAUAAAGUGUCCCAAAGUGGUCUGUCGAGACCCAUUUCGGACUAUGGCCACCUUGGCCAACAUAUCCACCA .................................((.(((((((((....)))).))))).))....(((((.....)))))............ (-11.56 = -11.68 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:04 2011