| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,116,251 – 14,116,341 |
| Length | 90 |
| Max. P | 0.993616 |

| Location | 14,116,251 – 14,116,341 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 80.57 |
| Shannon entropy | 0.35192 |
| G+C content | 0.38632 |
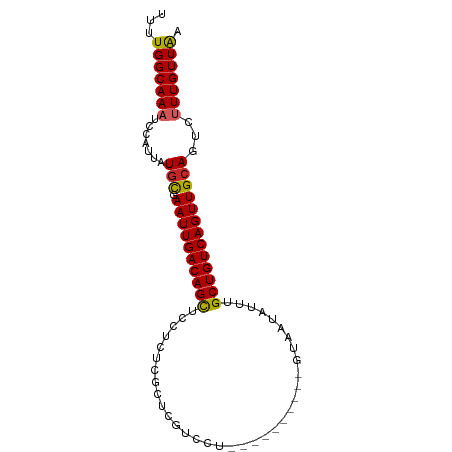
| Mean single sequence MFE | -18.90 |
| Consensus MFE | -16.55 |
| Energy contribution | -16.16 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
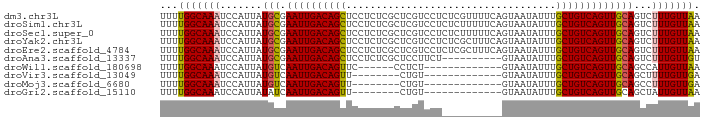
>dm3.chr3L 14116251 90 - 24543557 UUUUGGCAAAUCCAUUAUGCGAAUUGACAGCUCCUCUCGCUCGUCCUCUCGUUUUCAGUAAUAUUUGCUGUCAGUUGCAGUCUUUGUUAA ..((((((((..(....(((.((((((((((.......(((((......)).....))).......))))))))))))))..)))))))) ( -20.54, z-score = -2.66, R) >droSim1.chr3L 13471046 90 - 22553184 UUUUGGCAAAUCCAUUAUGCGAAUUGACAGCUCCUCUCGCUCGUCCUCUCUUUUUCAGUAAUAUUUGCUGUCAGUUGCAGUCUUUGUUAA ..((((((((..(....(((.((((((((((.......(((...............))).......))))))))))))))..)))))))) ( -19.70, z-score = -2.60, R) >droSec1.super_0 6263773 90 - 21120651 UUUUGGCAAAUCCAUUAUGCGAAUUGACAGCUCCUCUCGCUCGUCCUCUCUUUUUCAGUAAUAUUUGCUGUCAGUUGCAGUCUUUGUUAA ..((((((((..(....(((.((((((((((.......(((...............))).......))))))))))))))..)))))))) ( -19.70, z-score = -2.60, R) >droYak2.chr3L 14207572 90 - 24197627 UUUUGGCAAAUCCAUUAUGCGAAUUGACAGCUCCUCUCGCUCGUCCUCUCGCUUUCAGUAAUAUUUGCUGUCAGUUGCAGUCUUUGUUAA ..((((((((..(....(((.((((((((((.......(((((......)).....))).......))))))))))))))..)))))))) ( -20.54, z-score = -2.25, R) >droEre2.scaffold_4784 14106259 90 - 25762168 UUUUGGCAAAUCCAUUAUGCGAAUUGACAGCUCCUCUCGCUCGUCCUCUCGCUUUCAGUAAUAUUUGCUGUCAGUUGCAGUCUUUGUUAA ..((((((((..(....(((.((((((((((.......(((((......)).....))).......))))))))))))))..)))))))) ( -20.54, z-score = -2.25, R) >droAna3.scaffold_13337 18365453 80 + 23293914 UUUUGGCAAAUCCAUUAUGCGAAUUGACAGCUCCUCUCGCUCCUUCU----------GUAAUAUUUGCUGUCAGUUGCAGUCUUUGUUGU .....((((........(((.((((((((((.......((.......----------)).......))))))))))))).......)))) ( -19.50, z-score = -2.44, R) >droWil1.scaffold_180698 10006069 71 + 11422946 UUUUGGCAAAUCCAUUAUGUCAAUUGACAGUUC------CCUCU-------------GUAAUAUUUGCUGUCAGUUGCAGCCAUUGUUAA ...((((..........((.(((((((((((..------.....-------------.........)))))))))))))))))....... ( -17.59, z-score = -2.88, R) >droVir3.scaffold_13049 18390268 69 - 25233164 UUUUGGCAAAUCCAUUAUGUCAAUUGACAGUU--------CUGU-------------GUAAUAUUUGCUGUCAGUUGCAGCUUUUGUUGA ..(..(((((.......((.(((((((((((.--------....-------------.........)))))))))))))...)))))..) ( -18.94, z-score = -2.61, R) >droMoj3.scaffold_6680 19181857 69 - 24764193 UUUUGGCAAAUCCAUUAUGUCAAUUGACAGUU--------CUGU-------------GUAAUAUUUGCUGUCAGUUGCAGCCUUUGUUGA ..(..(((((.......((.(((((((((((.--------....-------------.........)))))))))))))...)))))..) ( -18.24, z-score = -2.35, R) >droGri2.scaffold_15110 15307198 69 + 24565398 UUUUGGCAAAUCCAUUAUAUCAAUUGACAGUU--------CUGU-------------GUAAUAUUUGCUGUCAGUUGCAGCUAUUGUUAA ..(((((((...........(((((((((((.--------....-------------.........)))))))))))......))))))) ( -13.67, z-score = -0.99, R) >consensus UUUUGGCAAAUCCAUUAUGCGAAUUGACAGCUCCUCUCGCUCGUCCU__________GUAAUAUUUGCUGUCAGUUGCAGUCUUUGUUAA ...(((((((.......(((.((((((((((...................................)))))))))))))...))))))). (-16.55 = -16.16 + -0.39)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:25:02 2011