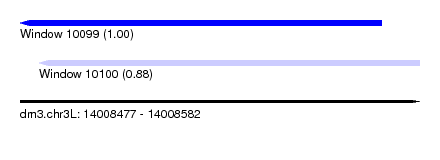
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,008,477 – 14,008,582 |
| Length | 105 |
| Max. P | 0.997714 |

| Location | 14,008,477 – 14,008,572 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 56.76 |
| Shannon entropy | 0.88471 |
| G+C content | 0.57723 |
| Mean single sequence MFE | -41.73 |
| Consensus MFE | -10.83 |
| Energy contribution | -9.13 |
| Covariance contribution | -1.70 |
| Combinations/Pair | 2.00 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.997714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14008477 95 - 24543557 ---AGGUGGUCGAGUCUUGAAUGCCACUUUGCUCACAUCGAGCAGAGUUGUCGCAGCGAUUGUUGCCGCAGUAGGUGUUGCAGUUACAGUCGCUGUCG ---.(((..((((...))))..)))(((((((((.....)))))))))....(((((((((((.((.((((......)))).)).))))))))))).. ( -39.40, z-score = -3.07, R) >droSim1.chr3L_random 671121 95 - 1049610 ---UGGUGGUCGAGUCUUGACUGCCACUUUGCUCACAUCGAGCAGAGUUGUCGCAGCGAUUGCUGCUGCAGUAGUUGUUGCAGUUGCAGUCGCUGUUG ---.((..(((((...)))))..))(((((((((.....)))))))))....(((((((((((.(((((((......))))))).))))))))))).. ( -51.50, z-score = -5.52, R) >droSec1.super_0 6154969 95 - 21120651 ---UGGUGGUCGAGUCUUGACUGCCACUUUGCUCACGUCGAGCAGAGUUGUCGCAGCGAUUGCUGCUGCAGUAGUUGUUGCAGUUACAGUCGCUGUCG ---.((..(((((...)))))..))(((((((((.....)))))))))....((((((((((..(((((((......)))))))..)))))))))).. ( -48.20, z-score = -4.64, R) >droYak2.chr3L 14096252 95 - 24197627 ---AGGUGGUUUAGUCUUCACUGCCGCUUUGCUAACAUCGAGCAGCGUGGCCGCAGCGACUGCUGCUGCAGUUGGUGUUGCAGUUACAGUUGCUGUCG ---.(((((........)))))(((((.(((((.......))))).))))).((((((((((..(((((((......)))))))..)))))))))).. ( -43.00, z-score = -2.61, R) >droEre2.scaffold_4784 14000475 95 - 25762168 ---AGGGGGUUUAGUCUUGACUGCCACUUUGCUCACAUCGAGCAGAGUUGUCGCAGCGACUGCUGCUGCAGUUGUUGAUGCAGUUGCAGUUGCUGUAG ---.((.((((.......)))).))(((((((((.....)))))))))....(((((((((((.((((((........)))))).))))))))))).. ( -48.40, z-score = -4.65, R) >droAna3.scaffold_13337 11358499 82 + 23293914 UGGCGGCGUCCUGGUUUUCAACAUCACCUUGCCCACAUCGAGGAGCGUGGCCGCCGCAGCAGCGGUCGCCGUGGCAGUCGCC---------------- .((((((.....(((..........))).((((.((..((.....)).(((.(((((....))))).)))))))))))))))---------------- ( -31.50, z-score = 0.25, R) >dp4.chrXR_group8 6783089 83 + 9212921 GGUUAGCGGACGGGAAUUGAUUGCACCUCCUGGUGCUGGUGCUACCGCUGUGGCUGUGGAUGUGGCUGCAGCUGUAGCUGUGC--------------- ..((((((..(((((..((....))..))))).)))))).(((((.((((..((..(....)..))..)))).))))).....--------------- ( -35.20, z-score = -1.98, R) >droPer1.super_24 624713 83 - 1556852 GGUUAGCGGACGGGAAUUGAUUGCACCUCCUGGUGCUGGUGCUACCGCUGUGGCUGUGGAUGUGGCUGCAGCUGUAGCUGUGC--------------- ..((((((..(((((..((....))..))))).)))))).(((((.((((..((..(....)..))..)))).))))).....--------------- ( -35.20, z-score = -1.98, R) >droMoj3.scaffold_6680 17721783 98 - 24764193 AGCUGCAGCGCGCGGUUUGGCAGUCGCCUUGGCCUCAACCACAGUCGCCGUUGCAAUGGCUGUUGUUGCUGUUGCUGGCAUUGCAGCCAUUGCUGCUG (((.((((((.(((..((((..(..((....))..)..))).)..)))))))))(((((((((...((((......))))..)))))))))))).... ( -43.20, z-score = -0.68, R) >consensus ___UGGUGGUCGAGUCUUGACUGCCACUUUGCUCACAUCGAGCAGCGUUGUCGCAGCGACUGCUGCUGCAGUUGUUGUUGCAGUU_CAGU_GCUGU_G ..............................................(((((.(((((....))))).))))).......................... (-10.83 = -9.13 + -1.70)
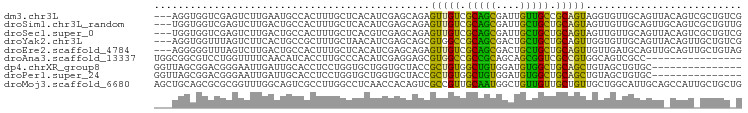
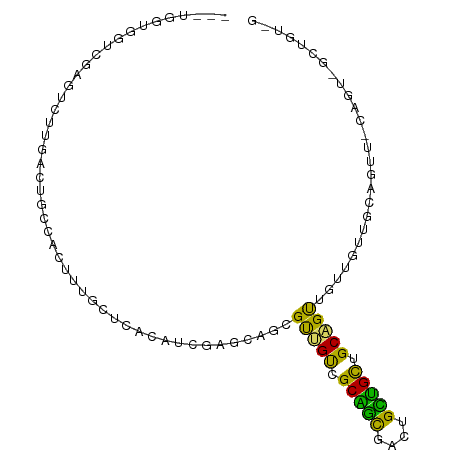
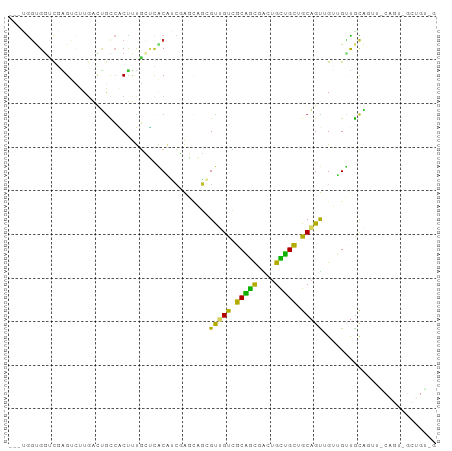
| Location | 14,008,482 – 14,008,582 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 58.04 |
| Shannon entropy | 0.86091 |
| G+C content | 0.56674 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -10.83 |
| Energy contribution | -9.13 |
| Covariance contribution | -1.70 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.881602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 14008482 100 - 24543557 CCAGUUUGAGAGGUGG---UCGAGUCUUGAAUGCCACUUUGCUCACAUCGAGCAGAGUUGUCGCAGCGAUUGUUGCCGCAGUAGGUGUUGCAGUUACAGUCGC ...(((..(((..(..---..)..)))..)))((.(((((((((.....))))))))).))....((((((((.((.((((......)))).)).)))))))) ( -34.50, z-score = -1.52, R) >droSim1.chr3L_random 671126 100 - 1049610 CCAGUUUGAGUGGUGG---UCGAGUCUUGACUGCCACUUUGCUCACAUCGAGCAGAGUUGUCGCAGCGAUUGCUGCUGCAGUAGUUGUUGCAGUUGCAGUCGC .((..((((((((..(---((((...)))))..))))))(((((.....)))))))..)).....((((((((.(((((((......))))))).)))))))) ( -45.60, z-score = -3.48, R) >droSec1.super_0 6154974 100 - 21120651 CCAGCUUGAGUGGUGG---UCGAGUCUUGACUGCCACUUUGCUCACGUCGAGCAGAGUUGUCGCAGCGAUUGCUGCUGCAGUAGUUGUUGCAGUUACAGUCGC .((((((((((((..(---((((...)))))..))))))(((((.....))))))))))).....(((((((..(((((((......)))))))..))))))) ( -47.40, z-score = -3.75, R) >droYak2.chr3L 14096257 100 - 24197627 CCAGUUUAACAGGUGG---UUUAGUCUUCACUGCCGCUUUGCUAACAUCGAGCAGCGUGGCCGCAGCGACUGCUGCUGCAGUUGGUGUUGCAGUUACAGUUGC ...........(((((---........)))))(((((.(((((.......))))).)))))....(((((((..(((((((......)))))))..))))))) ( -36.60, z-score = -0.59, R) >droEre2.scaffold_4784 14000480 100 - 25762168 CCAGCUUAAGAGGGGG---UUUAGUCUUGACUGCCACUUUGCUCACAUCGAGCAGAGUUGUCGCAGCGACUGCUGCUGCAGUUGUUGAUGCAGUUGCAGUUGC ...((......((.((---((.......)))).))(((((((((.....)))))))))....)).((((((((.((((((........)))))).)))))))) ( -43.60, z-score = -2.70, R) >droAna3.scaffold_13337 11358500 91 + 23293914 CUGCCUUGAAUGGCGGCGUCCUGGUUUUCAACAUCACCUUGCCCACAUCGAGGAGCGUGGCCGCCGCAGCAGCGGUCGCCGUGGCAGUCGC------------ (((((.((...(((.(((((((.(.......((......)).......).)))).))).)))(((((....)))))...)).)))))....------------ ( -33.14, z-score = 0.67, R) >dp4.chrXR_group8 6783094 88 + 9212921 UCAUCUUGAGGGUUAGCGGACGGGAAUUGAUUGCACCUCCUGGUGCUGGUGCUACCGCUGUGGCUGUGGAUGUGGCUGCAGCUGUAGC--------------- ............((((((..(((((..((....))..))))).)))))).(((((.((((..((..(....)..))..)))).)))))--------------- ( -34.00, z-score = -1.24, R) >droPer1.super_24 624718 88 - 1556852 UCAUCUUGAGGGUUAGCGGACGGGAAUUGAUUGCACCUCCUGGUGCUGGUGCUACCGCUGUGGCUGUGGAUGUGGCUGCAGCUGUAGC--------------- ............((((((..(((((..((....))..))))).)))))).(((((.((((..((..(....)..))..)))).)))))--------------- ( -34.00, z-score = -1.24, R) >droMoj3.scaffold_6680 17721788 103 - 24764193 CAUCCUUCACAGCUGCAGCGCGCGGUUUGGCAGUCGCCUUGGCCUCAACCACAGUCGCCGUUGCAAUGGCUGUUGUUGCUGUUGCUGGCAUUGCAGCCAUUGC ...........((.((((((.(((..((((..(..((....))..)..))).)..)))))))))(((((((((...((((......))))..))))))))))) ( -41.10, z-score = -0.65, R) >consensus CCAGCUUGAGAGGUGG___UCGAGUCUUGACUGCCACUUUGCUCACAUCGAGCAGCGUUGUCGCAGCGACUGCUGCUGCAGUUGUUGUUGCAGUU_CAGU_GC ........................................................(((((.(((((....))))).)))))..................... (-10.83 = -9.13 + -1.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:58 2011