| Sequence ID | dm3.chr3L |
|---|---|
| Location | 14,005,798 – 14,005,889 |
| Length | 91 |
| Max. P | 0.821072 |

| Location | 14,005,798 – 14,005,889 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 75.07 |
| Shannon entropy | 0.52057 |
| G+C content | 0.60121 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -10.81 |
| Energy contribution | -10.18 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.69 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.821072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
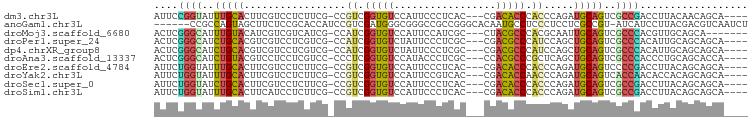
>dm3.chr3L 14005798 91 + 24543557 AUUCCGGUAUUUGCACUUCGUCCUCUUCG-CCGUCGGUGUCCAUUCCCUCAC---CGACACCCACCCAGAUGCAGUCGCCGACCUUACAACAGCA---- ....((((..(((((((.((.......))-..(((((((..........)))---))))........)).)))))..))))..............---- ( -22.00, z-score = -2.62, R) >anoGam1.chr3L 38516658 92 + 41284009 ------CCGCCAGUAGCUUCUCCGCACCAUCCGUCGAUGGGCGGGCCGCCGGGCACAAUGCCUCCCUCCUCGCCGU-AUCAUCCUUACGACGUCAAUCU ------..(((.(..((....((((.(((((....)))))))))))..)..)))................((.(((-(.......)))).))....... ( -25.40, z-score = 0.02, R) >droMoj3.scaffold_6680 17719038 88 + 24764193 ACUCGGGCAUUUGUACAUCGUCGUCAUCG-CCAUCGGUGUCCAUUCCAUCGC---CUACGCCCACGCAAUUGCAGUCGCCCACGUUGCAGCA------- ....((((...((.((((((.((....))-....)))))).)).......(.---...)))))..((...(((((.((....))))))))).------- ( -20.00, z-score = 0.19, R) >droPer1.super_24 621836 91 + 1556852 ACUCGGGCAUCUGCACGUCGUCCUCGUCG-CCAUCGGUGUCUAUUCCCUCGC---CGACGCCCAUCCAGCUGCAGUCGCCCACAUUGCAGCAGCA---- ....((((....(((((.......))).)-)..((((((..........)))---))).)))).....((((((((.......))))))))....---- ( -30.60, z-score = -2.45, R) >dp4.chrXR_group8 6780311 91 - 9212921 ACUCGGGCAUCUGCACGUCGUCCUCGUCG-CCAUCGGUGUCUAUUCCCUCGC---CGACGCCCAUCCAGCUGCAGUCGCCCACAUUGCAGCAGCA---- ....((((....(((((.......))).)-)..((((((..........)))---))).)))).....((((((((.......))))))))....---- ( -30.60, z-score = -2.45, R) >droAna3.scaffold_13337 11355915 91 - 23293914 ACUCGGGCAUCUGUACGUCCUCCUCGUCC-CCCUCGGUGUCCAUACCCUCGC---CCACGCCCGCUCAGCUGCAGUCGCCCACCCUGCAGCACCA---- ...(((((......(((.......)))..-.....((((..........)))---)...)))))....(((((((.........)))))))....---- ( -25.30, z-score = -2.60, R) >droEre2.scaffold_4784 13997820 91 + 25762168 AUUCUGGUAUUUGCACUUCGUCCUCUUCG-CCGUCGGUGUCCAUUCCCUCAC---CGACACCCACCCAGAUGCAGUCCCCGACCUUACAGCAGCA---- ...((((((((((.....((.......))-..(((((((..........)))---)))).......))))))).(((...)))....))).....---- ( -18.30, z-score = -1.24, R) >droYak2.chr3L 14093582 91 + 24197627 AUUCUGGUAUUUGCACUUCGUCCUCUUCG-CCGUCGGUGUCCAUUCCGUCAC---CGACACCAACCCAGAUGCAGUCACCAACACCACAGCAGCA---- ....((((..(((((((.((.......))-..(((((((.(......).)))---))))........)).)))))..))))..............---- ( -21.60, z-score = -2.23, R) >droSec1.super_0 6152272 91 + 21120651 AUUCUGGUAUCUGCACUUCGUCCUCUUCG-CCGUCGGUGUCCAUUCCCUCAC---CGACACCCACCCAGAUGCAGUCGCCGACCUUACAGCAGCA---- ....((((..(((((((.((.......))-..(((((((..........)))---))))........)).)))))..))))..............---- ( -22.10, z-score = -1.85, R) >droSim1.chr3L 13361029 91 + 22553184 AUUCUGGUAUUUGCACUUCAUCCUCUUCG-CCGUCGGUGUCCAUUCCCUCAC---CGACACCCACCCAGAUGCAGUCGCCGACCUUACAGCAGCA---- .....(((....(((((.((((.......-..(((((((..........)))---)))).........)))).))).))..)))...........---- ( -21.07, z-score = -2.11, R) >consensus AUUCGGGCAUCUGCACUUCGUCCUCUUCG_CCGUCGGUGUCCAUUCCCUCAC___CGACACCCACCCAGAUGCAGUCGCCCACCUUACAGCAGCA____ ....((((..(((((.................((.(((((.................))))).)).....)))))..)))).................. (-10.81 = -10.18 + -0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:57 2011