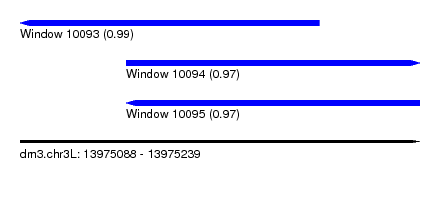
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,975,088 – 13,975,239 |
| Length | 151 |
| Max. P | 0.985802 |

| Location | 13,975,088 – 13,975,201 |
|---|---|
| Length | 113 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.29 |
| Shannon entropy | 0.53692 |
| G+C content | 0.64172 |
| Mean single sequence MFE | -44.87 |
| Consensus MFE | -24.21 |
| Energy contribution | -25.38 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.985802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13975088 113 - 24543557 -CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAU--CACCUGCAAAUU-GGCGCACUGCCCACGCCGGCGAGUGAAACGCCACGAUUUG -((..((((.......))))((((....(((.((((....)))).)))(((((((..((((.--....))))..))-)))))........))))((((.......)))).))..... ( -46.90, z-score = -1.93, R) >anoGam1.chr2L 42778566 101 + 48795086 ----------------CACGGCCGACCCGUUUGUCAAUCACUGCUACAGUUACGAUCUGCACGACAGCAUCGAGCUGGGCGCGUUGCCGACACCGGCGAGCGAGGUGGCUAACGUUG ----------------...((((.(((((((((((...........(((((.(((((((.....))).)))))))))(((.....)))......)))))))).)))))))....... ( -38.00, z-score = -0.98, R) >droGri2.scaffold_15110 6170630 86 + 24565398 ----------------------------CAGUGGCACUGGCGGCGGUGGUGCCGAGAUGCAU--CAAUUGCAAAUU-GGCGCCCUGCCCACGCCGGCGAGUGAAACGCCCAGAUUUG ----------------------------..(.(((.((((((((((.((((((((..((((.--....))))..))-)))))))))))...)))))...(.....)))))....... ( -39.50, z-score = -2.34, R) >droVir3.scaffold_13049 2100772 86 + 25233164 ----------------------------CAGCGGCACUGGCGGUGUCGGUGCCGAGCUGCAU--CAUUUGCAAAUU-GGCGCCCUGCCGACGCCGGCGAGUGAAACGCCAAGAUUUG ----------------------------..(((.((((.((((((((((((((((..((((.--....))))..))-))).....))))))))).)).))))...)))......... ( -42.30, z-score = -3.03, R) >droAna3.scaffold_13337 11327245 113 + 23293914 -CGAUGCUGACACCAUUGGCGGCGACUCCAGCGGCUGCUAUGCCACUGGCGCCGAGCUGCAG--CAUCUGCAAAUU-GGCGCACUGCCCACGCCGGCGAGUGAAACGCCCCGAUUUG -.(((((((.((...((((((......((((.(((......))).))))))))))..)))))--))))..((((((-(((((((((((......))).))))...))))..)))))) ( -48.40, z-score = -2.15, R) >droEre2.scaffold_4784 13965022 113 - 25762168 -CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAU--CACCUGCAAAUU-GGCGCACUGCCCACGCCGGCGAGUGAAACGCCACGAUUUG -((..((((.......))))((((....(((.((((....)))).)))(((((((..((((.--....))))..))-)))))........))))((((.......)))).))..... ( -46.90, z-score = -1.93, R) >droYak2.chr3L 14062234 114 - 24197627 ACGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAU--CACCUGCAAAUU-GGCGCACUGCCCACGCCGGCGAGUGAAACGCCACGAUUUG .((..((((.......))))((((....(((.((((....)))).)))(((((((..((((.--....))))..))-)))))........))))((((.......)))).))..... ( -47.40, z-score = -2.04, R) >droSec1.super_0 6121929 113 - 21120651 -CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGUUGCAU--CACCUGCAAAUU-GGCGCACUGCCCACGCCGGCGAGUGAAACGCCACGAUUUG -((..((((.......))))((((....(((.((((....)))).)))(((((((.(((((.--....))))).))-)))))........))))((((.......)))).))..... ( -47.50, z-score = -2.10, R) >droSim1.chr3L 13331456 113 - 22553184 -CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAU--CACCUGCAAAUU-GGCGCACUGCCCACGCCGGCGAGUGAAACGCCACGAUUUG -((..((((.......))))((((....(((.((((....)))).)))(((((((..((((.--....))))..))-)))))........))))((((.......)))).))..... ( -46.90, z-score = -1.93, R) >consensus _CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAU__CACCUGCAAAUU_GGCGCACUGCCCACGCCGGCGAGUGAAACGCCACGAUUUG ....................((((....(((.(((......))).)))(((((((..((((.......))))..)).)))))((((((......))).)))....))))........ (-24.21 = -25.38 + 1.17)

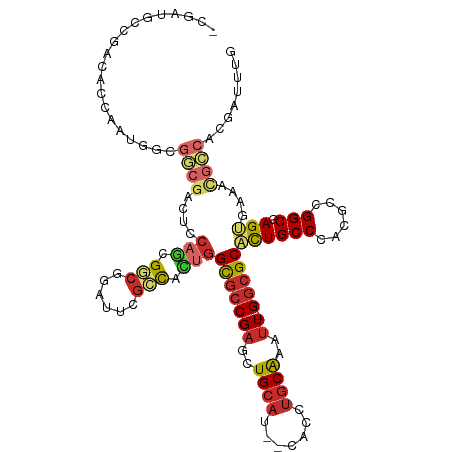
| Location | 13,975,128 – 13,975,239 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 96.60 |
| Shannon entropy | 0.05721 |
| G+C content | 0.62142 |
| Mean single sequence MFE | -48.14 |
| Consensus MFE | -44.90 |
| Energy contribution | -44.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13975128 111 + 24543557 AAUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG---CCGUAACCACCAACCACACCGACAUGGCCAUAGCCGUA .....(..((((........((((((((((((((....)))))))))...)))))...((((((.((((...)---)))....))))))...))))..)(((((....))))). ( -49.00, z-score = -2.41, R) >droEre2.scaffold_4784 13965062 108 + 25762168 AAUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG---CCGUAACCACCAACCACACCGACAUGGCCAUAGCC--- ....((..((((........((((((((((((((....)))))))))...)))))...((((((.((((...)---)))....))))))...))))..)).(((....)))--- ( -46.40, z-score = -2.19, R) >droYak2.chr3L 14062274 111 + 24197627 AAUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCGUAACCGUAACCACCAACCACACCGACAUGGCCGUAGCC--- ....((((.....))))(((((((.(((((((((....))))))))).((((......((((((.(((........)))....))))))......))))...)))).))).--- ( -47.30, z-score = -2.22, R) >droSec1.super_0 6121969 111 + 21120651 AAUUUGCAGGUGAUGCAACUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG---CCGUAACCACCAACCACACCGACAUGGCCAUAGCCGUA .....(..((((........((((((((((((((....)))))))))...)))))...((((((.((((...)---)))....))))))...))))..)(((((....))))). ( -49.00, z-score = -2.76, R) >droSim1.chr3L 13331496 111 + 22553184 AAUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG---CCGUAACCACCAACCACACCGACAUGGCCAUAGCCGUA .....(..((((........((((((((((((((....)))))))))...)))))...((((((.((((...)---)))....))))))...))))..)(((((....))))). ( -49.00, z-score = -2.41, R) >consensus AAUUUGCAGGUGAUGCAGCUCGGCGCCAGUGGCGAAUCCGCCGCUGGAGUCGCCGCCAUUGGUGUCGGCAUCG___CCGUAACCACCAACCACACCGACAUGGCCAUAGCCGUA ....((..((((........((((((((((((((....)))))))))...)))))...((((((.(((........)))....))))))...))))..)).(((....)))... (-44.90 = -44.90 + -0.00)

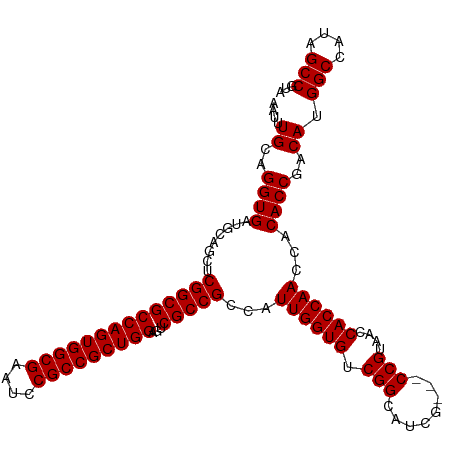
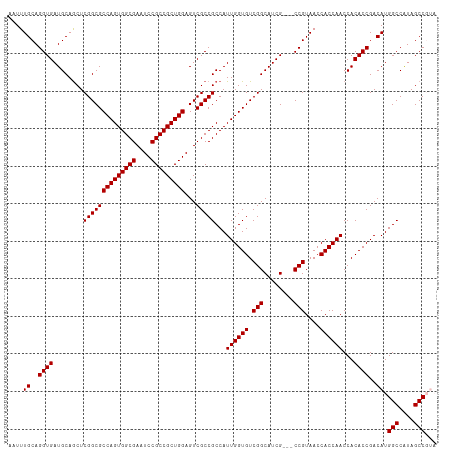
| Location | 13,975,128 – 13,975,239 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 96.60 |
| Shannon entropy | 0.05721 |
| G+C content | 0.62142 |
| Mean single sequence MFE | -52.08 |
| Consensus MFE | -49.16 |
| Energy contribution | -48.84 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.968221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13975128 111 - 24543557 UACGGCUAUGGCCAUGUCGGUGUGGUUGGUGGUUACGG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAUU ...((((.((((...((((.(((.(((((((....(((---(...)))).))))))).))).)))).((((.((((....)))).)))).))))))))(((.....)))..... ( -51.90, z-score = -2.15, R) >droEre2.scaffold_4784 13965062 108 - 25762168 ---GGCUAUGGCCAUGUCGGUGUGGUUGGUGGUUACGG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAUU ---((((.((((...((((.(((.(((((((....(((---(...)))).))))))).))).)))).((((.((((....)))).)))).))))))))(((.....)))..... ( -51.60, z-score = -2.22, R) >droYak2.chr3L 14062274 111 - 24197627 ---GGCUACGGCCAUGUCGGUGUGGUUGGUGGUUACGGUUACGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAUU ---((((.((((...((((.(((.(((((((....((((......)))).))))))).))).)))).((((.((((....)))).)))).))))))))(((.....)))..... ( -53.90, z-score = -2.92, R) >droSec1.super_0 6121969 111 - 21120651 UACGGCUAUGGCCAUGUCGGUGUGGUUGGUGGUUACGG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGUUGCAUCACCUGCAAAUU ..((((.........((((.(((.(((((((....(((---(...)))).))))))).))).)))).((((.((((....)))).)))).))))..(((((.....)))))... ( -51.10, z-score = -2.08, R) >droSim1.chr3L 13331496 111 - 22553184 UACGGCUAUGGCCAUGUCGGUGUGGUUGGUGGUUACGG---CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAUU ...((((.((((...((((.(((.(((((((....(((---(...)))).))))))).))).)))).((((.((((....)))).)))).))))))))(((.....)))..... ( -51.90, z-score = -2.15, R) >consensus UACGGCUAUGGCCAUGUCGGUGUGGUUGGUGGUUACGG___CGAUGCCGACACCAAUGGCGGCGACUCCAGCGGCGGAUUCGCCACUGGCGCCGAGCUGCAUCACCUGCAAAUU ...((((.((((...((((.(((.(((((((....(((........))).))))))).))).)))).((((.((((....)))).)))).))))))))(((.....)))..... (-49.16 = -48.84 + -0.32)

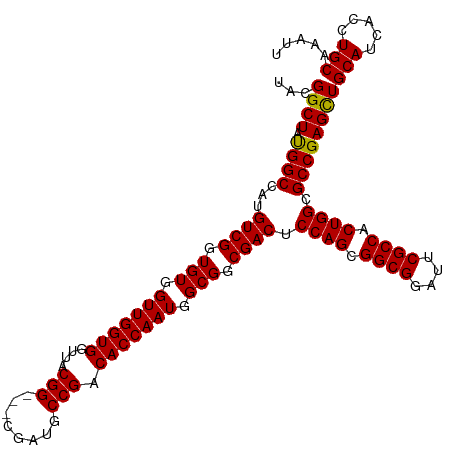
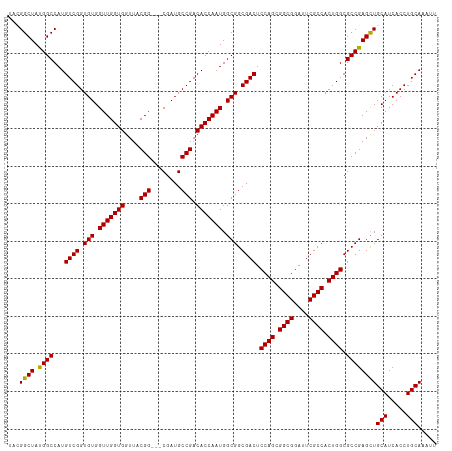
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:54 2011