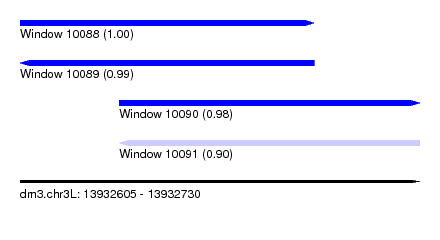
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,932,605 – 13,932,730 |
| Length | 125 |
| Max. P | 0.997808 |

| Location | 13,932,605 – 13,932,697 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 71.24 |
| Shannon entropy | 0.57937 |
| G+C content | 0.36046 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -13.62 |
| Energy contribution | -13.55 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.997808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
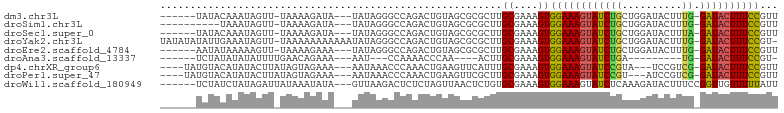
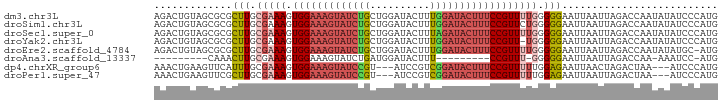
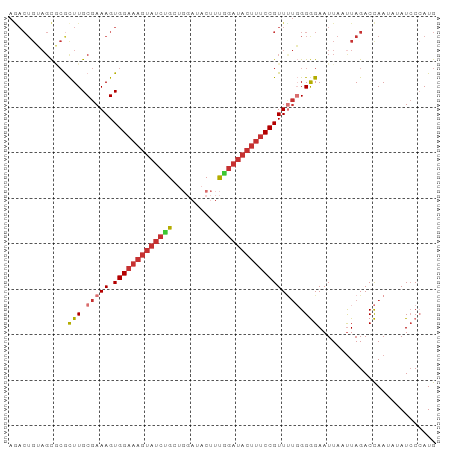
>dm3.chr3L 13932605 92 + 24543557 ------UAUACAAAUAGUU-UAAAAGAUA---UAUAGGGCCAGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUG-GAUACUUUCCGUU ------.............-...(((...---(((((.......))))).....)))((....))((((((((((((..(....)..))-))))))))))... ( -22.50, z-score = -1.15, R) >droSim1.chr3L 13288892 88 + 22553184 ----------UAAAUAGUU-UAAAAGAUA---UAUAGGGCCAGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUG-GAUACUUUCCGUU ----------.........-...(((...---(((((.......))))).....)))((....))((((((((((((..(....)..))-))))))))))... ( -22.50, z-score = -1.33, R) >droSec1.super_0 6079694 92 + 21120651 ------UAUACAAAUAGUU-UAAAAGAUA---UAUAGGGCCAGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUA-GAUACUUUCCGUU ------.............-...(((...---(((((.......))))).....)))((....))((((((((((((..(....)..))-))))))))))... ( -23.80, z-score = -1.73, R) >droYak2.chr3L 14019575 100 + 24197627 UAUAUAUAUUGAAAUAGUU-UAAAAAAAAAAAUAUAGGGCCAGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUG-GAUACUUUCCGU- ...................-............(((((.......))))).(((....))).....((((((((((((..(....)..))-))))))))))..- ( -21.50, z-score = -1.19, R) >droEre2.scaffold_4784 13923816 92 + 25762168 ------AAUAUAAAAAGUU-UAAAAGAAA---UAUAGGGCCAGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUG-GAUACUUUCCGUU ------.............-...(((...---(((((.......))))).....)))((....))((((((((((((..(....)..))-))))))))))... ( -22.50, z-score = -1.50, R) >droAna3.scaffold_13337 11282502 76 - 23293914 ------UCUAUAUAUAUUUUGAACAGAAA---AAU---CCAAAACCCAA----ACUUGCGAAAGUGGAAAGUAUCUGA---------UG-GAUACUUUCCGU- ------.......................---...---...........----....((....))((((((((((...---------..-))))))))))..- ( -17.00, z-score = -3.23, R) >dp4.chrXR_group6 5754946 92 + 13314419 ----UAUGUACAUAUACUUAUAGUAGAAA---AAUAAACCCAAACUGAAGUUCAUUUGCGAAAGUGGAAAGUAUCCGUA---UCCGUCG-GAUACUUUCCGUU ----..((((.((..((((.((((.....---...........))))))))..)).)))).....((((((((((((..---.....))-))))))))))... ( -24.09, z-score = -3.88, R) >droPer1.super_47 246576 92 + 592741 ----UAUGUACAUAUACUUAUAGUAGAAA---AAUAAACCCAAACUGAAGUUCGCUUGCGAAAGUGGAAAGUAUCCGU---AUCCGUCG-GAUACUUUCCGUU ----..((((.....((((.((((.....---...........)))))))).....)))).....((((((((((((.---......))-))))))))))... ( -23.39, z-score = -3.24, R) >droWil1.scaffold_180949 3545704 94 - 6375548 ------UCUAUCUAUAGAUUAUAAAUAUA---GUUAAGACUCUCUAGUUAACUCUGUGCGAAAGUGGAAAGUAUCUCAAAGAUACUUUCCGGUUGUUUUUAUU ------(((..(((((.........))))---)...))).........(((((....((....))((((((((((.....)))))))))))))))........ ( -21.40, z-score = -2.25, R) >consensus ______UAUACAAAUAGUU_UAAAAGAAA___UAUAGGGCCAGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUG_GAUACUUUCCGUU .........................................................((....))((((((((((...............))))))))))... (-13.62 = -13.55 + -0.07)
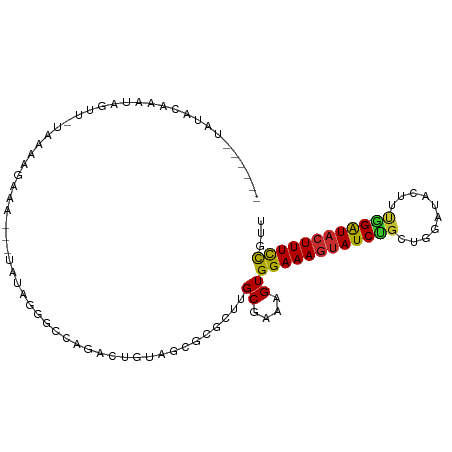
| Location | 13,932,605 – 13,932,697 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 71.24 |
| Shannon entropy | 0.57937 |
| G+C content | 0.36046 |
| Mean single sequence MFE | -19.59 |
| Consensus MFE | -9.43 |
| Energy contribution | -9.98 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.994199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
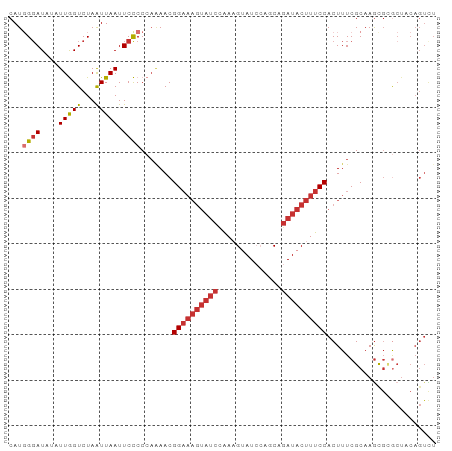
>dm3.chr3L 13932605 92 - 24543557 AACGGAAAGUAUC-CAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCUGGCCCUAUA---UAUCUUUUA-AACUAUUUGUAUA------ ...((((((((((-..............))))))))))......((....))((((.....))))......---.........-.............------ ( -16.24, z-score = -0.92, R) >droSim1.chr3L 13288892 88 - 22553184 AACGGAAAGUAUC-CAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCUGGCCCUAUA---UAUCUUUUA-AACUAUUUA---------- ...((((((((((-..............))))))))))......((....))((((.....))))......---.........-.........---------- ( -16.24, z-score = -1.44, R) >droSec1.super_0 6079694 92 - 21120651 AACGGAAAGUAUC-UAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCUGGCCCUAUA---UAUCUUUUA-AACUAUUUGUAUA------ ...((((((((((-(...((.....)))))))))))))......((....))((((.....))))......---.........-.............------ ( -17.90, z-score = -1.35, R) >droYak2.chr3L 14019575 100 - 24197627 -ACGGAAAGUAUC-CAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCUGGCCCUAUAUUUUUUUUUUUA-AACUAUUUCAAUAUAUAUA -..((((((((((-..............))))))))))......((....))((((.....))))..................-................... ( -16.24, z-score = -1.55, R) >droEre2.scaffold_4784 13923816 92 - 25762168 AACGGAAAGUAUC-CAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCUGGCCCUAUA---UUUCUUUUA-AACUUUUUAUAUU------ ...((((((((((-..............))))))))))......((....))((((.....))))......---.........-.............------ ( -16.24, z-score = -1.55, R) >droAna3.scaffold_13337 11282502 76 + 23293914 -ACGGAAAGUAUC-CA---------UCAGAUACUUUCCACUUUCGCAAGU----UUGGGUUUUGG---AUU---UUUCUGUUCAAAAUAUAUAUAGA------ -..((((((((((-..---------...)))))))))).((..(....).----..))(((((((---((.---.....))))))))).........------ ( -22.30, z-score = -4.40, R) >dp4.chrXR_group6 5754946 92 - 13314419 AACGGAAAGUAUC-CGACGGA---UACGGAUACUUUCCACUUUCGCAAAUGAACUUCAGUUUGGGUUUAUU---UUUCUACUAUAAGUAUAUGUACAUA---- ...((((((((((-((.....---..))))))))))))........((((((((((......)))))))))---)...........(((....)))...---- ( -25.30, z-score = -3.66, R) >droPer1.super_47 246576 92 - 592741 AACGGAAAGUAUC-CGACGGAU---ACGGAUACUUUCCACUUUCGCAAGCGAACUUCAGUUUGGGUUUAUU---UUUCUACUAUAAGUAUAUGUACAUA---- ...((((((((((-((......---.)))))))))))).....(.(((((........))))).)......---............(((....)))...---- ( -25.00, z-score = -3.31, R) >droWil1.scaffold_180949 3545704 94 + 6375548 AAUAAAAACAACCGGAAAGUAUCUUUGAGAUACUUUCCACUUUCGCACAGAGUUAACUAGAGAGUCUUAAC---UAUAUUUAUAAUCUAUAGAUAGA------ .............((((((((((.....))))))))))((((((...............))))))......---..((((((((...))))))))..------ ( -20.86, z-score = -3.02, R) >consensus AACGGAAAGUAUC_CAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCUGGCCCUAUA___UUUCUUUUA_AACUAUUUAUAUA______ ...((((((((((...............))))))))))................................................................. ( -9.43 = -9.98 + 0.56)

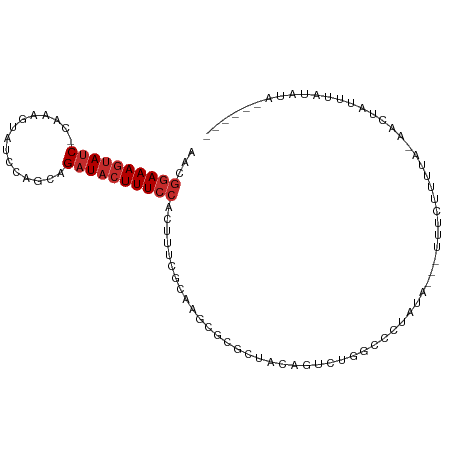
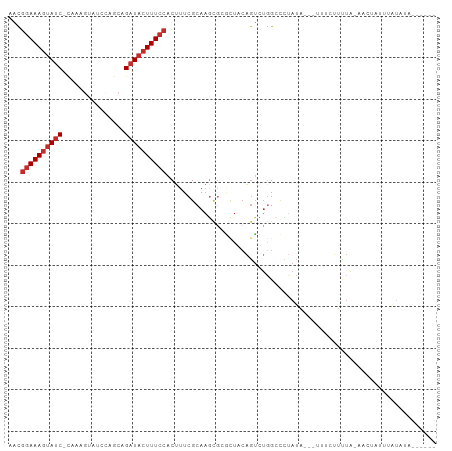
| Location | 13,932,636 – 13,932,730 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 83.45 |
| Shannon entropy | 0.33830 |
| G+C content | 0.41171 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -17.39 |
| Energy contribution | -19.03 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
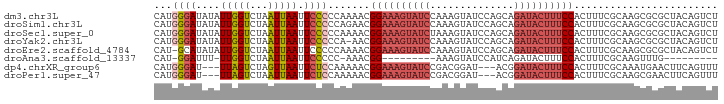
>dm3.chr3L 13932636 94 + 24543557 AGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUGGAUACUUUCCGUUUUGGGGGAAUUAAUUAGACCAAUAUAUCCCAUG ..(((....(((....)))..)))((((((((((((..(....)..))))))))))))....(((((....................))))).. ( -24.85, z-score = -1.36, R) >droSim1.chr3L 13288919 94 + 22553184 AGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUGGAUACUUUCCGUUCUGGGGGAAUUAAUUAGACCAAUAUAUCCCAUG ...((..(((......((....))((((((((((((..(....)..)))))))))))))))..))((((..................))))... ( -24.87, z-score = -1.19, R) >droSec1.super_0 6079725 94 + 21120651 AGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUAGAUACUUUCCGUUUUGGGGGAAUUAAUUAGACCAAUAUAUCCCAUG ..(((....(((....)))..)))((((((((((((..(....)..))))))))))))....(((((....................))))).. ( -26.15, z-score = -2.01, R) >droYak2.chr3L 14019615 93 + 24197627 AGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUGGAUACUUUCCGUU-UGGGGGAAUUAAUUAGACCAAUAUAUCCCAUG ..(((....(((....)))..)))((((((((((((..(....)..))))))))))))...-(((((....................))))).. ( -24.85, z-score = -1.43, R) >droEre2.scaffold_4784 13923847 93 + 25762168 AGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUGGAUACUUUCCGUUUUGGGGGAAUUAAUUAGACCAAUAUAUGC-AUG ...(((.......(((.(((((.(((((((((((((..(....)..)))))))))))))))))).))).......)))............-... ( -24.34, z-score = -1.55, R) >droAna3.scaffold_13337 11282536 73 - 23293914 ---------CAAACUUGCGAAAGUGGAAAGUAUCUGAUGGAUACUUU---------CCGUUU-GGGGGAAUUAAUUAGACCAA-AAAUCC-AUG ---------(((((..((....))((((((((((.....))))))))---------))))))-).((............))..-......-... ( -22.00, z-score = -3.51, R) >dp4.chrXR_group6 5754980 88 + 13314419 AAACUGAAGUUCAUUUGCGAAAGUGGAAAGUAUCCGU---AUCCGUCGGAUACUUUCCGUUUUUGGAGAAUUAACUAGACUAA---AUCCCAUG .......((((.((((.(((((((((((((((((((.---......)))))))))))))))))))..)))).)))).......---........ ( -28.80, z-score = -4.68, R) >droPer1.super_47 246610 88 + 592741 AAACUGAAGUUCGCUUGCGAAAGUGGAAAGUAUCCGU---AUCCGUCGGAUACUUUCCGUUUUUGGAGAAUUAAUUAGACUAA---AUCCCAUG ...(((((((((.....(((((((((((((((((((.---......)))))))))))))))))))..)))))..)))).....---........ ( -28.00, z-score = -4.18, R) >consensus AGACUGUAGCGCGCUUGCGAAAGUGGAAAGUAUCUGCUGGAUACUUUGGAUACUUUCCGUUUUGGGGGAAUUAAUUAGACCAAUAUAUCCCAUG ...(((.......(((.(((((.(((((((((((((..(....)..)))))))))))))))))).))).......)))................ (-17.39 = -19.03 + 1.64)
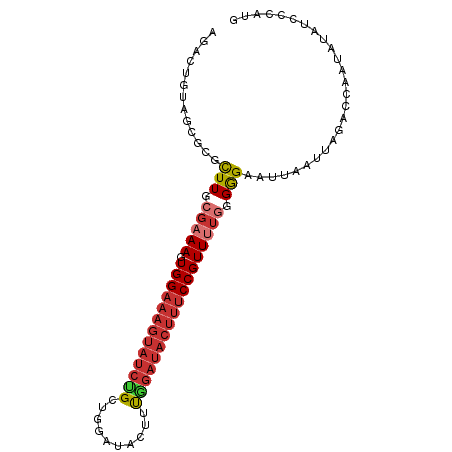
| Location | 13,932,636 – 13,932,730 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 83.45 |
| Shannon entropy | 0.33830 |
| G+C content | 0.41171 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -11.48 |
| Energy contribution | -12.67 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
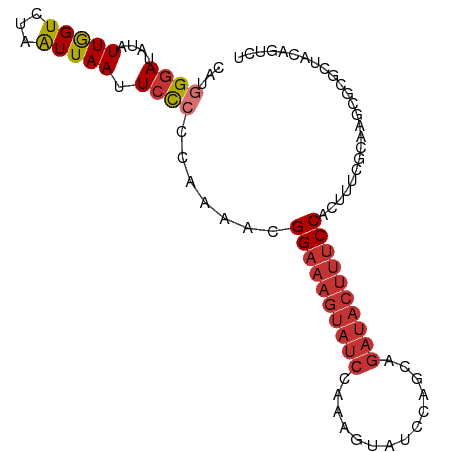
>dm3.chr3L 13932636 94 - 24543557 CAUGGGAUAUAUUGGUCUAAUUAAUUCCCCCAAAACGGAAAGUAUCCAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCU ..((((....((((((...))))))...))))....((((((((((..............)))))))))).....(((....)))......... ( -20.24, z-score = -1.76, R) >droSim1.chr3L 13288919 94 - 22553184 CAUGGGAUAUAUUGGUCUAAUUAAUUCCCCCAGAACGGAAAGUAUCCAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCU ..((((....((((((...))))))...))))....((((((((((..............)))))))))).....(((....)))......... ( -20.24, z-score = -1.33, R) >droSec1.super_0 6079725 94 - 21120651 CAUGGGAUAUAUUGGUCUAAUUAAUUCCCCCAAAACGGAAAGUAUCUAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCU ..((((....((((((...))))))...))))....(((((((((((...((.....))))))))))))).....(((....)))......... ( -21.90, z-score = -2.28, R) >droYak2.chr3L 14019615 93 - 24197627 CAUGGGAUAUAUUGGUCUAAUUAAUUCCCCCA-AACGGAAAGUAUCCAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCU ..((((....((((((...))))))...))))-...((((((((((..............)))))))))).....(((....)))......... ( -20.24, z-score = -1.77, R) >droEre2.scaffold_4784 13923847 93 - 25762168 CAU-GCAUAUAUUGGUCUAAUUAAUUCCCCCAAAACGGAAAGUAUCCAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCU ..(-((.....((((..............))))...((((((((((..............))))))))))......)))............... ( -17.28, z-score = -1.96, R) >droAna3.scaffold_13337 11282536 73 + 23293914 CAU-GGAUUU-UUGGUCUAAUUAAUUCCCCC-AAACGG---------AAAGUAUCCAUCAGAUACUUUCCACUUUCGCAAGUUUG--------- ..(-((((..-...)))))...........(-((((((---------((((((((.....)))))))))).(....)...)))))--------- ( -16.90, z-score = -2.94, R) >dp4.chrXR_group6 5754980 88 - 13314419 CAUGGGAU---UUAGUCUAGUUAAUUCUCCAAAAACGGAAAGUAUCCGACGGAU---ACGGAUACUUUCCACUUUCGCAAAUGAACUUCAGUUU ..(((((.---((((.....)))).)).))).((((((((((((((((......---.))))))))))))...((((....)))).....)))) ( -24.30, z-score = -3.15, R) >droPer1.super_47 246610 88 - 592741 CAUGGGAU---UUAGUCUAAUUAAUUCUCCAAAAACGGAAAGUAUCCGACGGAU---ACGGAUACUUUCCACUUUCGCAAGCGAACUUCAGUUU ..(((((.---(((((...))))).)).))).((((((((((((((((......---.))))))))))))...((((....)))).....)))) ( -28.00, z-score = -4.66, R) >consensus CAUGGGAUAUAUUGGUCUAAUUAAUUCCCCCAAAACGGAAAGUAUCCAAAGUAUCCAGCAGAUACUUUCCACUUUCGCAAGCGCGCUACAGUCU ...((((..................)))).......((((((((((..............))))))))))........................ (-11.48 = -12.67 + 1.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:51 2011