
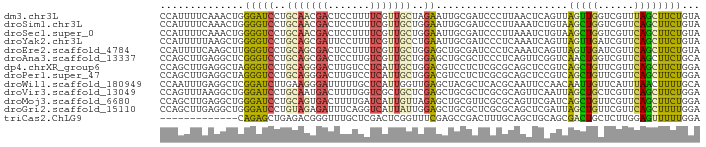
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,925,134 – 13,925,224 |
| Length | 90 |
| Max. P | 0.701604 |

| Location | 13,925,134 – 13,925,224 |
|---|---|
| Length | 90 |
| Sequences | 13 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 69.63 |
| Shannon entropy | 0.69782 |
| G+C content | 0.51390 |
| Mean single sequence MFE | -29.59 |
| Consensus MFE | -8.79 |
| Energy contribution | -8.30 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.88 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13925134 90 - 24543557 CCAUUUUCAAACUGGGAUCCUGCAACGACUCCUUUUCGUUGCUAGAAUUGCGAUCCCUUAACUCAGUUAGUUGGUCGUUUAGCUUCUGUA .............(((((((((((((((.......))))))).))......))))))......(((..((((((....)))))).))).. ( -22.50, z-score = -2.15, R) >droSim1.chr3L 13281474 90 - 22553184 CCAUUUUCAAACUGGGGUCCUGCAACGACUCCUUUUCGUUGCUGGAAUUGCGAUCCCUUAAAUCUGUAAGCUGGUCGUUCAGCUUCUGUA .............(((((((.(((((((.......))))))).))).......))))..........(((((((....)))))))..... ( -24.11, z-score = -2.10, R) >droSec1.super_0 6072713 90 - 21120651 CCAUUUUCAAACUGGGGUCCUGCAACGACUCCUUUUCGUUGCUGGAAUUGCGAUCCCUUAAAUCUGUAAGCUGGUCGUUCAGCUUCUGUA .............(((((((.(((((((.......))))))).))).......))))..........(((((((....)))))))..... ( -24.11, z-score = -2.10, R) >droYak2.chr3L 14011377 90 - 24197627 CCAUUUUUAAGCUGGGGUCCUGCAGCGAUUCCUUUUCGUUGCCUGAAUUGCGAUCCCUCAAAUCAGUUAGUUGAUCGUUCAGCUUCUGUA .............(((((((.(((((((.......))))))).......).))))))......(((..((((((....)))))).))).. ( -24.70, z-score = -2.26, R) >droEre2.scaffold_4784 13916592 90 - 25762168 CCAUUUUCAAGCUUGGGUCCUGCAGCGACUCCUUUUCGUUGCUGGAGCUGCGAUCCCUCAAAUCAGUUAGUUGAUCGUUCAGCUUCUGUA ..........((..((.(((.(((((((.......))))))).))).))))............(((..((((((....)))))).))).. ( -26.20, z-score = -1.79, R) >droAna3.scaffold_13337 11273442 90 + 23293914 CCAGCUUGAGGCUCGGGUCCUGCAGCGACUCCUUGUCGUUGCUGGAGCUGCGCUCCCUCAGUUCGGUCAACUGGUCGUUCAGCUUCUGCA ...((..(((((((((.(((.((((((((.....)))))))).))).)))..........(..((..(....)..))..))))))).)). ( -37.40, z-score = -1.88, R) >dp4.chrXR_group6 5747365 90 - 13314419 CCAGCUUGAGGCUAGGGUCCUGCAGGGACUUGUCCUCAUUGCUGGACGUCCUCUCGCGCAGCUCCGUCAGCUGUUCGUUCAGCUUCUGGA ((((..(((((...((((((.....))))))..)))))..((((((((.........((((((.....)))))).))))))))..)))). ( -36.90, z-score = -1.51, R) >droPer1.super_47 239006 90 - 592741 CCAGCUUGAGGCUAGGGUCCUGCAGGGACUUGUCCUCAUUGCUGGACGUCCUCUCGCGCAGCUCCGUCAGCUGUUCGUUCAGCUUCUGGA ((((..(((((...((((((.....))))))..)))))..((((((((.........((((((.....)))))).))))))))..)))). ( -36.90, z-score = -1.51, R) >droWil1.scaffold_180949 3535321 90 + 6375548 CCAAUUUGAGGCUCGGAUCUUGAAGGGAUUUUUGCUCAUUGGUUGAGCUACGCUCACGCAAUUCCAACAAUUGUUCAUUUAACUUUUGCA ....((..(((......)))..))((((((...(((((.....)))))...((....))))))))..(((..(((.....)))..))).. ( -17.20, z-score = 0.30, R) >droVir3.scaffold_13049 24049013 90 - 25233164 CCAGUUUAAGGCUGGGAUCCUGCAAUGACUUUUGGUCGCUGCUCGAGCUGCGCUCGCGCAGUUCAAUUAGCUGCUCGUUCAGCUUCUGGA ((((....((((((((.....(((..(((.....)))((((...((((((((....))))))))...)))))))...)))))))))))). ( -36.00, z-score = -2.09, R) >droMoj3.scaffold_6680 18673817 90 - 24764193 CCAGCUUGAGGCUGGGAUCCUGCAGUGACUUUUGAUCAUUGUUAGAGCUGCGUUCGCGCAGUUCGAUCAGCUGUUCGUUCAGCUUCUGGA (((((.....)))))((((..((((((((....).)))))))..((((((((....))))))))))))(((((......)))))...... ( -37.90, z-score = -2.97, R) >droGri2.scaffold_15110 14909385 90 + 24565398 CCAGCUUGAGGCUGGGAUCCUGUAGAGAUUUCAGGUCAUUAUUGGAGCUGCGCUCGCGCAGCUCGAUUAGCUGUUCGUUCAGCUUUUGGA (((((.....)))))((.((((.........)))))).......((((((((....))))))))....(((((......)))))...... ( -35.50, z-score = -2.03, R) >triCas2.ChLG9 6521147 77 - 15222296 -------------CAGAGCUGAGACGGGUUUGCUCGACUCGGUUUCGAGCCGACUUUGCAGCUGCAGCGACUGCUCUUGGAGUUUUUGGA -------------...(((((....(((((.((((((.......)))))).)))))..))))).(((.((((.(....).)))).))).. ( -25.20, z-score = 0.09, R) >consensus CCAGUUUGAGGCUGGGGUCCUGCAGCGACUUCUUUUCGUUGCUGGAGCUGCGAUCCCGCAGCUCAGUCAGCUGUUCGUUCAGCUUCUGGA ..............(((((..(((((((.......)))))))..))......................(((((......))))))))... ( -8.79 = -8.30 + -0.49)
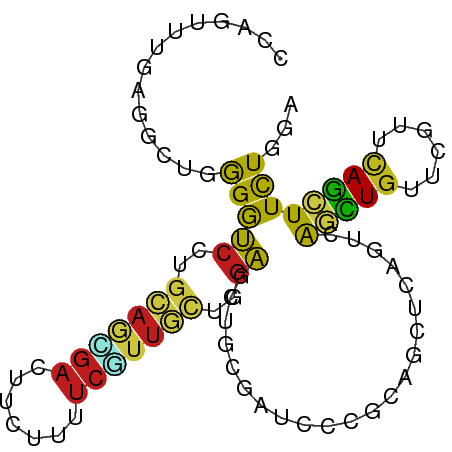

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:48 2011