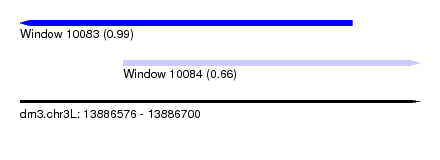
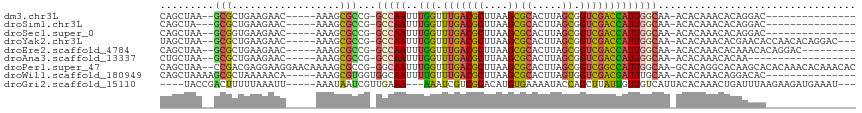
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,886,576 – 13,886,700 |
| Length | 124 |
| Max. P | 0.990793 |

| Location | 13,886,576 – 13,886,679 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.46 |
| Shannon entropy | 0.65237 |
| G+C content | 0.48815 |
| Mean single sequence MFE | -27.09 |
| Consensus MFE | -14.52 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.990793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
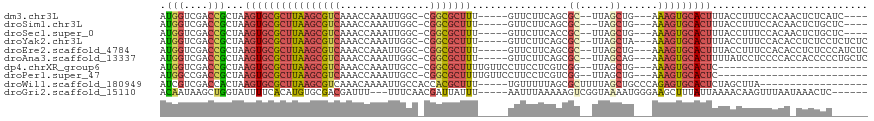
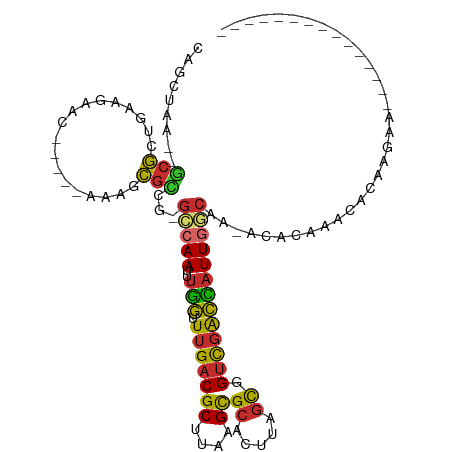
>dm3.chr3L 13886576 103 - 24543557 AUGGUCGACCGCUAAGUGCGCUUAAGCGUCAAACCAAAUUGGC-CGGCGCUUU-----GUUCUUCAGCGC--UUAGCUG---AAAGUGCACUUUACCUUUCCACAACUCUCAUC---- .(((.........(((((((((((((((((...((.....)).-.))))))).-----.....(((((..--...))))---))))))))))).......)))...........---- ( -32.79, z-score = -2.94, R) >droSim1.chr3L 13242084 102 - 22553184 AUGGUCGACCGCUAAGUGCGCUUAAGCGUCAAACCAAAUUGGC-CGGCGCUUU-----GUUCUUCAGCGC---UAGCUG---AAAGUGCACUUUACCUUUCCACAACUCUGCUC---- .(((.........(((((((((((((((((...((.....)).-.))))))).-----.....(((((..---..))))---))))))))))).......)))...........---- ( -32.79, z-score = -2.29, R) >droSec1.super_0 6028942 103 - 21120651 AUGGUCGACCGCUAAGUGCGCUUAAGCGUCAAACCAAAUUGGC-CGGCGCUUU-----GUUCUUCACCGC--UUAGCUG---AAAGUGCACUUUACCUUUCCACAACUCUGCUC---- .(((.........(((((((((((((((((...((.....)).-.))))))).-----.....(((.(..--...).))---))))))))))).......)))...........---- ( -26.19, z-score = -0.80, R) >droYak2.chr3L 13970605 107 - 24197627 AUGGUCGACCGCUAAGUGCGCUUAAGCGUCAAACCAAAUUGGC-CGGCGCUUU-----GUUCUUCAGCGC--UUAGCUA---AAAGUGCACUUUACCUUUCCACACCUCUCCUCUCUC .(((.........((((((((((.((((((((......)))))-.((((((..-----.......)))))--)..))).---.)))))))))).......)))............... ( -28.89, z-score = -2.36, R) >droEre2.scaffold_4784 13876702 107 - 25762168 AUGGUCGACCGCUAAGUGCGCUUAAGCGUCAAACCAAAUUGGC-CGGCGCUUU-----GUUCUUCAGCGC--UUAGCUG---AAAGUGCACUUUACCUUUCCACACCUCUCCCAUCUC .(((.........(((((((((((((((((...((.....)).-.))))))).-----.....(((((..--...))))---))))))))))).......)))............... ( -32.79, z-score = -3.10, R) >droAna3.scaffold_13337 11238256 107 + 23293914 AUGGUCGACCGCUAAGUGCGCUUAAGCGUCAAACCAAAUUGGC-CGGCGCUUU-----GUUCUUCAGCGC--UUAGCAG---AAAGUGCACUUUUAUCCUCCCCACCACCCCCUGCUC .((((.(...(((((((((....(((((((...((.....)).-.))))))).-----........))))--))))).(---((((....))))).......).)))).......... ( -30.52, z-score = -1.90, R) >dp4.chrXR_group6 5706121 87 - 13314419 AUGGUCGACCGCUAAGUGCGCUUAAGCGUCAAACCAAAUUGCC-CGGCGCUUUUGUUCCUUCCUCGUCGG--UUAGCUG---AAAGUGCACUC------------------------- ..((....))....((((((((((((((((.............-.)))))))..............((((--....)))---)))))))))).------------------------- ( -24.04, z-score = -0.81, R) >droPer1.super_47 198592 87 - 592741 AUGGCCGACCGCUAAGUGCGCUUAAGCGUCAAACCAAAUUGCC-CGGCGCUUUUGUUCCUUCCUCGUCGG--UUAGCUG---AAAGUGCACUC------------------------- .((((.....))))((((((((((((((((.............-.)))))))..............((((--....)))---)))))))))).------------------------- ( -25.14, z-score = -0.93, R) >droWil1.scaffold_180949 3479729 95 + 6375548 AUCGUCGACCACUAAGUGCGCUUAAGCGUCAAACAAAAUUGCCACCACGCUUU-----UGUUUUUAGCGCUUUUAGCUGCCCAGAGUGCACUCUAGCUUA------------------ ...........((((((((((((((((((.((((((((..((......)))))-----)))))...))))))...........))))))))).)))....------------------ ( -24.10, z-score = -1.27, R) >droGri2.scaffold_15110 9210922 104 + 24565398 ACAAUAAGCUGGUAUUUUCACAUGUGCGACGAUUU---UUUCAACGAUUAUUU-----AAUUUAAAAAGUCGGUAAAAUGGGAAGCUUUAUUAAAACAAGUUUAAUAAACUC------ ......((((.((((........))))..(.((((---(.....(((((.(((-----(...)))).)))))..))))).)..))))((((((((.....))))))))....------ ( -13.70, z-score = 0.17, R) >consensus AUGGUCGACCGCUAAGUGCGCUUAAGCGUCAAACCAAAUUGGC_CGGCGCUUU_____GUUCUUCAGCGC__UUAGCUG___AAAGUGCACUUUACCUUUCCACA_CUCUCC______ .(((....)))...((((((((((((((((...............)))))))................((.....))......))))))))).......................... (-14.52 = -15.20 + 0.68)
| Location | 13,886,608 – 13,886,700 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.57 |
| Shannon entropy | 0.57096 |
| G+C content | 0.48459 |
| Mean single sequence MFE | -24.89 |
| Consensus MFE | -13.17 |
| Energy contribution | -12.86 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664872 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
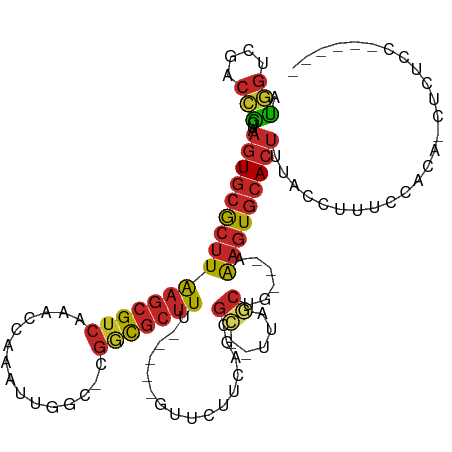
>dm3.chr3L 13886608 92 + 24543557 CAGCUAA--GCGCUGAAGAAC-----AAAGCGCCG-GCCAAUUUGGUUUGACGCUUAAGCGCACUUAGCGGUCGACCAUUGGCAA-ACACAAACACAGGAC--------------- .......--(((((.......-----..)))))..-(((((..(((((.((((((.(((....)))))).)))))))))))))..-...............--------------- ( -26.10, z-score = -1.10, R) >droSim1.chr3L 13242116 91 + 22553184 CAGCUA---GCGCUGAAGAAC-----AAAGCGCCG-GCCAAUUUGGUUUGACGCUUAAGCGCACUUAGCGGUCGACCAUUGGCAA-ACACAAACACAGGAC--------------- ..((..---(((((.......-----..)))))..-))...(((((((((.((((.(((....)))))))(((((...)))))..-...))))).))))..--------------- ( -26.50, z-score = -1.25, R) >droSec1.super_0 6028974 92 + 21120651 CAGCUAA--GCGGUGAAGAAC-----AAAGCGCCG-GCCAAUUUGGUUUGACGCUUAAGCGCACUUAGCGGUCGACCAUUGGCAA-ACACAAACACAGGAC--------------- .......--.(((((......-----....)))))-(((((..(((((.((((((.(((....)))))).)))))))))))))..-...............--------------- ( -24.70, z-score = -0.60, R) >droYak2.chr3L 13970641 104 + 24197627 UAGCUAA--GCGCUGAAGAAC-----AAAGCGCCG-GCCAAUUUGGUUUGACGCUUAAGCGCACUUAGCGGUCGACCAUUGGCAA-ACACAAACACGAACACCAACACAGGAC--- .......--(((((.......-----..)))))..-(((((..(((((.((((((.(((....)))))).)))))))))))))..-...............((......))..--- ( -27.10, z-score = -1.32, R) >droEre2.scaffold_4784 13876738 98 + 25762168 CAGCUAA--GCGCUGAAGAAC-----AAAGCGCCG-GCCAAUUUGGUUUGACGCUUAAGCGCACUUAGCGGUCGACCAUUGGCAA-ACACAAACACAAACACAGGAC--------- .......--(((((.......-----..)))))..-(((((..(((((.((((((.(((....)))))).)))))))))))))..-.....................--------- ( -26.10, z-score = -1.18, R) >droAna3.scaffold_13337 11238292 89 - 23293914 CUGCUAA--GCGCUGAAGAAC-----AAAGCGCCG-GCCAAUUUGGUUUGACGCUUAAGCGCACUUAGCGGUCGACCAUUGGCAA-ACACAAACACAA------------------ .......--(((((.......-----..)))))..-(((((..(((((.((((((.(((....)))))).)))))))))))))..-............------------------ ( -26.10, z-score = -1.06, R) >droPer1.super_47 198603 112 + 592741 CAGCUAA--CCGACGAGGAAGGAACAAAAGCGCCG-GGCAAUUUGGUUUGACGCUUAAGCGCACUUAGCGGUCGGCCAUUGGCAA-GCACAGGCACAAGCACACAAACACAAACAC ..(((((--(((((.............(((((.((-(((......))))).)))))...(((.....))))))))...)))))..-((....))...................... ( -27.10, z-score = -0.20, R) >droWil1.scaffold_180949 3479750 95 - 6375548 CAGCUAAAAGCGCUAAAAACA-----AAAGCGUGGUGGCAAUUUUGUUUGACGCUUAAGCGCACUUAGUGGUCGACGAUUUGCAA-ACACAAACAGGACAC--------------- ..((((...(((((.......-----..)))))..))))..(((((((((..((....))(((....((.....))....)))..-...)))))))))...--------------- ( -23.70, z-score = -0.58, R) >droGri2.scaffold_15110 9210959 101 - 24565398 ----UACCGACUUUUUAAAUU-----AAAUAAUCGUUGAAA---AAAUCGUCGCACAUGUGAAAAUACCAGCUUAUUGUUGUCAUUACACAAACUGAUUUAAGAAGAUGAAAU--- ----...((.(((((((((((-----(......((.(((..---...))).))....((((..(((..((((.....))))..))).))))...)))))))))))).))....--- ( -16.60, z-score = -1.40, R) >consensus CAGCUAA__GCGCUGAAGAAC_____AAAGCGCCG_GCCAAUUUGGUUUGACGCUUAAGCGCACUUAGCGGUCGACCAUUGGCAA_ACACAAACACAAGAA_______________ .........(((..................)))...(((((..(((.(((((((....))((.....)).)))))))))))))................................. (-13.17 = -12.86 + -0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:45 2011