| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,845,559 – 13,845,611 |
| Length | 52 |
| Max. P | 0.914858 |

| Location | 13,845,559 – 13,845,611 |
|---|---|
| Length | 52 |
| Sequences | 7 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 65.90 |
| Shannon entropy | 0.64296 |
| G+C content | 0.40744 |
| Mean single sequence MFE | -12.27 |
| Consensus MFE | -4.81 |
| Energy contribution | -5.10 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
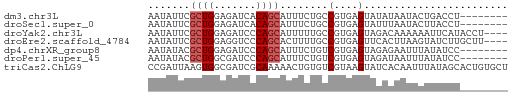
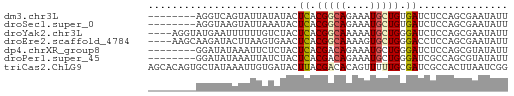
>dm3.chr3L 13845559 52 + 24543557 AAUAUUCGCUGGAGAUCACAGCAUUUCUGCCGUGAGUAUAUAAUACUGACCU-------- .((((((((.(.(((..........))).).)))))))).............-------- ( -10.90, z-score = -1.31, R) >droSec1.super_0 5988721 52 + 21120651 AAUAUUCGCUGGAGAUCACAGCAUUUCUGCCGUGAGUAUUUAAUACUUACCU-------- .......((((.......)))).........((((((((...))))))))..-------- ( -12.20, z-score = -2.42, R) >droYak2.chr3L 13928298 56 + 24197627 AAUAUUCGCUGGAGAUCCCAGCAUUUUUGCCGUGAGUAGACAAAAAAUUCAUACCU---- .......(((((.....))))).((((((((....)..).))))))..........---- ( -11.50, z-score = -2.05, R) >droEre2.scaffold_4784 13836095 56 + 25762168 AAUAUUCGCUGGAGGUCCCAGCACUUUUGCCGUGAGUUCACUUAAGUAUCUUGCUU---- .......(((((.....))))).........(..((...((....))..))..)..---- ( -12.30, z-score = -0.87, R) >dp4.chrXR_group8 6391997 52 + 9212921 AAUAUACGCUGGAGAUCCCAGCAUUUCUGUCGUGAGUAGAGAAUUUAUAUCC-------- .(((((.(((((.....))))).((((((.(....)))))))...)))))..-------- ( -15.40, z-score = -3.37, R) >droPer1.super_45 528089 52 - 618639 AAUAUACGCUGGCGAUCCCAGCAUUUCUGUCGUGAGUAGAUAAUUUAUAUCC-------- .(((((.(((((.....)))))...((((.(....))))).....)))))..-------- ( -12.40, z-score = -2.08, R) >triCas2.ChLG9 9473649 60 + 15222296 CCGAUUAAGUGGCGAUCGCAAAAACUGUGUCGUAAGUAUCACAAUUUAUAGCACUGUGCU ..((((.......))))(((......((((..(((((......)))))..))))..))). ( -11.20, z-score = -0.15, R) >consensus AAUAUUCGCUGGAGAUCCCAGCAUUUCUGCCGUGAGUAGAUAAUAUUUACCU________ .......((((.......))))...................................... ( -4.81 = -5.10 + 0.29)
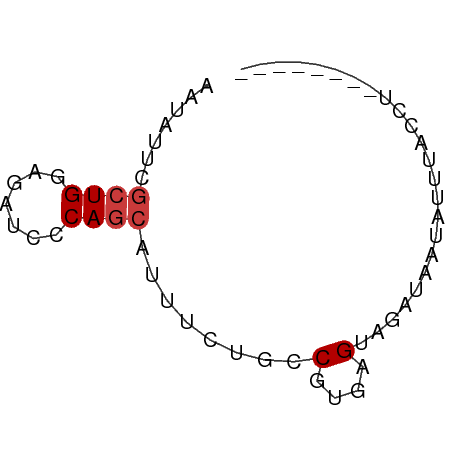
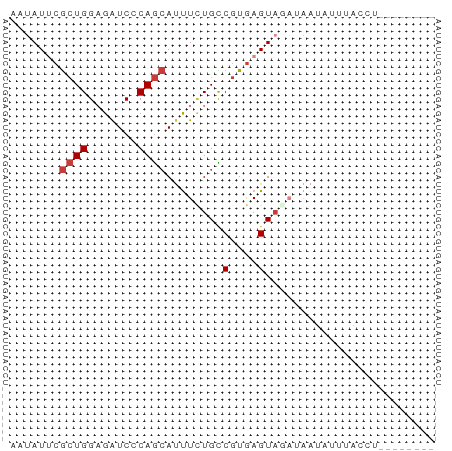
| Location | 13,845,559 – 13,845,611 |
|---|---|
| Length | 52 |
| Sequences | 7 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 65.90 |
| Shannon entropy | 0.64296 |
| G+C content | 0.40744 |
| Mean single sequence MFE | -12.40 |
| Consensus MFE | -4.60 |
| Energy contribution | -4.74 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13845559 52 - 24543557 --------AGGUCAGUAUUAUAUACUCACGGCAGAAAUGCUGUGAUCUCCAGCGAAUAUU --------.((...((((...))))((((((((....))))))))...)).......... ( -14.50, z-score = -2.46, R) >droSec1.super_0 5988721 52 - 21120651 --------AGGUAAGUAUUAAAUACUCACGGCAGAAAUGCUGUGAUCUCCAGCGAAUAUU --------.((...((((...))))((((((((....))))))))...)).......... ( -14.50, z-score = -2.95, R) >droYak2.chr3L 13928298 56 - 24197627 ----AGGUAUGAAUUUUUUGUCUACUCACGGCAAAAAUGCUGGGAUCUCCAGCGAAUAUU ----..........((((((((.......))))))))((((((.....))))))...... ( -14.00, z-score = -2.03, R) >droEre2.scaffold_4784 13836095 56 - 25762168 ----AAGCAAGAUACUUAAGUGAACUCACGGCAAAAGUGCUGGGACCUCCAGCGAAUAUU ----.........((((..(((....))).....))))(((((.....)))))....... ( -11.30, z-score = -0.66, R) >dp4.chrXR_group8 6391997 52 - 9212921 --------GGAUAUAAAUUCUCUACUCACGACAGAAAUGCUGGGAUCUCCAGCGUAUAUU --------............................(((((((.....)))))))..... ( -9.80, z-score = -1.13, R) >droPer1.super_45 528089 52 + 618639 --------GGAUAUAAAUUAUCUACUCACGACAGAAAUGCUGGGAUCGCCAGCGUAUAUU --------(((((.....))))).............(((((((.....)))))))..... ( -12.60, z-score = -2.30, R) >triCas2.ChLG9 9473649 60 - 15222296 AGCACAGUGCUAUAAAUUGUGAUACUUACGACACAGUUUUUGCGAUCGCCACUUAAUCGG .((....(((...((((((((..........))))))))..)))...))........... ( -10.10, z-score = -0.28, R) >consensus ________AGGUAAAUAUUAUAUACUCACGGCAGAAAUGCUGGGAUCUCCAGCGAAUAUU .........................((.(((((....))))).))............... ( -4.60 = -4.74 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:39 2011