
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,202,230 – 5,202,285 |
| Length | 55 |
| Max. P | 0.723961 |

| Location | 5,202,230 – 5,202,285 |
|---|---|
| Length | 55 |
| Sequences | 14 |
| Columns | 62 |
| Reading direction | forward |
| Mean pairwise identity | 72.18 |
| Shannon entropy | 0.60456 |
| G+C content | 0.56738 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -5.89 |
| Energy contribution | -6.45 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 2.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5202230 55 + 23011544 AGAGCUCACGUUGCGCUUCGAUACACUGC-AGUCGCAGUGGAAGGCGCGACAGCAG------ ...(((...(((((((((.....((((((-....))))))..))))))))))))..------ ( -25.00, z-score = -2.95, R) >droSim1.chr2L 4999465 55 + 22036055 AGAGCUCACGUUGCGCUUCGAUACACUGC-AAUCGCAGUGGAAGGCGCGACAGCAG------ ...(((...(((((((((.....((((((-....))))))..))))))))))))..------ ( -25.00, z-score = -3.48, R) >droSec1.super_5 3283431 55 + 5866729 AGAGCUCACGUUGCGCUUCGAUACACUGC-AAUCGCAGUGGAAGGCGCGACAGCAA------ ...(((...(((((((((.....((((((-....))))))..))))))))))))..------ ( -25.00, z-score = -3.51, R) >droYak2.chr2L 11999017 55 - 22324452 AGAGCUCACGUUGCGCUUCGAUACACUGC-AAUCGCAGUGGAAGGCGCGACAGCAG------ ...(((...(((((((((.....((((((-....))))))..))))))))))))..------ ( -25.00, z-score = -3.48, R) >droEre2.scaffold_4929 5280259 55 + 26641161 AGAGCUCACGUUGCGCUUCGAUACACUGC-AAUCGCAGUGGAAGGCGCGACAGCAG------ ...(((...(((((((((.....((((((-....))))))..))))))))))))..------ ( -25.00, z-score = -3.48, R) >droAna3.scaffold_12916 6217326 55 - 16180835 AGAGCUCACGUUGCGCUUUGACACACUGC-AGUCGCAGUGGAAGGCGCGACAGCAG------ ...(((...((((((((((....((((((-....)))))).)))))))))))))..------ ( -26.30, z-score = -3.22, R) >dp4.chr4_group3 9001658 55 - 11692001 AGAGCUCACGUUGCGCUUUGACACACUGC-AAUCGCAGUGGAAGGCGCGACAGCAG------ ...(((...((((((((((....((((((-....)))))).)))))))))))))..------ ( -26.30, z-score = -3.90, R) >droPer1.super_1 6101703 55 - 10282868 AGAGCUCACGUUGCGCUUUGACACACUGC-AAUCGCAGUGGAAGGCGCGACAGCAG------ ...(((...((((((((((....((((((-....)))))).)))))))))))))..------ ( -26.30, z-score = -3.90, R) >droWil1.scaffold_180772 7827406 55 - 8906247 AGAGCUGGCGUUGCGCUUUGAUACACUGC-AAUCGCAGUGGAAAGCGCGACAGCAG------ .......((((((((((((....((((((-....)))))).)))))))))).))..------ ( -26.60, z-score = -3.73, R) >droMoj3.scaffold_6500 6810196 55 - 32352404 AGAGCUAGCGUUGCGCUUCGAUACCCUGC-AGUCGCAGUGGAAGGCGCGCCAACAG------ .........(((((((.((....((((((-....)))).))..)).))).))))..------ ( -17.40, z-score = 0.08, R) >droVir3.scaffold_12963 13808511 55 - 20206255 AGAGCUGGCGCUGCGCUUCGAUACGCUGC-AGUCGCAGUGGAAGGCGCGCCAACAG------ ...(.((((((.((.((((....((((((-....)))))))))))))))))).)..------ ( -26.10, z-score = -1.61, R) >anoGam1.chr3R 16650773 52 - 53272125 AGAAACGGAACGAU---AUCAUCGUUUGUUGACCGU-GUGGAAGGAGAAGCAAGAA------ .......(((((((---...)))))))(((..((..-......))...))).....------ ( -7.50, z-score = 0.42, R) >apiMel3.Group4 9662960 61 - 10796202 GGAGAUCAGAUUGUCCAAGUACAGGGUGUUGGCGGU-GCACACGGAGAAGCUGCUCCAGCAG ((((..(((.((.(((..((((...((....)).))-))....))).)).)))))))..... ( -15.10, z-score = 0.60, R) >triCas2.chrUn_44 243911 55 + 525920 UUACCUCAUCCAACGUUUCGAAACACUUCUAGUAACAGCCAAGGACA-AAGAACAG------ ........(((...(((....)))((.....)).........)))..-........------ ( -1.90, z-score = 1.16, R) >consensus AGAGCUCACGUUGCGCUUCGAUACACUGC_AAUCGCAGUGGAAGGCGCGACAGCAG______ .........(((.(((((......(((((.....)))))...))))).)))........... ( -5.89 = -6.45 + 0.56)
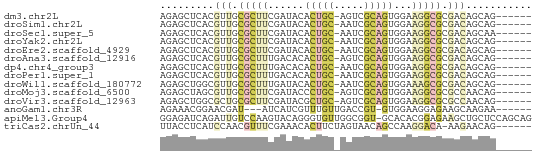

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:17 2011