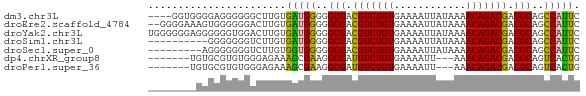
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,792,997 – 13,793,065 |
| Length | 68 |
| Max. P | 0.997915 |

| Location | 13,792,997 – 13,793,065 |
|---|---|
| Length | 68 |
| Sequences | 7 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 75.40 |
| Shannon entropy | 0.44939 |
| G+C content | 0.53070 |
| Mean single sequence MFE | -18.20 |
| Consensus MFE | -17.79 |
| Energy contribution | -17.29 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.21 |
| Mean z-score | -4.92 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.997431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
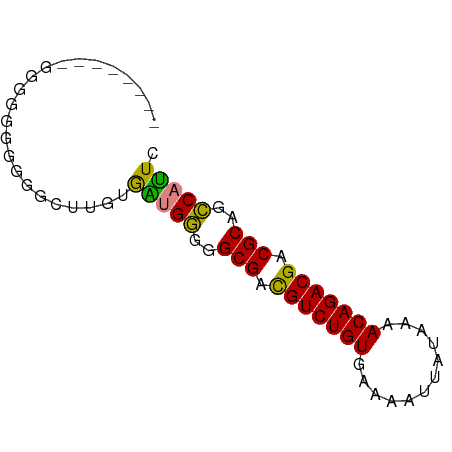
>dm3.chr3L 13792997 68 + 24543557 GAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGCCCCCCUCCCCACC---- ((.(((..(((.(((((((............))))))).)))..)))))...................---- ( -18.90, z-score = -5.42, R) >droEre2.scaffold_4784 13799886 70 + 25762168 GAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGUCCCCCCACUUUCCCC-- ((.(((..(((.(((((((............))))))).)))..))))).....................-- ( -18.90, z-score = -5.62, R) >droYak2.chr3L 13893283 72 + 24197627 GAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGUCCACCCCCCUCCCCCCA ((.(((..(((.(((((((............))))))).)))..)))))....................... ( -18.90, z-score = -5.75, R) >droSim1.chr3L 13179058 62 + 22553184 GAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGACCCCCCC---------- ((.(((..(((.(((((((............))))))).)))..))))).............---------- ( -18.90, z-score = -5.15, R) >droSec1.super_0 5954675 63 + 21120651 GAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCACCACAAGACCCCCCCU--------- ...(((..(((.(((((((............))))))).)))..)))................--------- ( -17.80, z-score = -4.74, R) >dp4.chrXR_group8 2643976 62 - 9212921 CAGUGACUGCGUCGUCUGUUU---AAUUUUCACAGACAUCGCCUUCGCUUUCUCCCACACGCACA------- .(((((..(((..((((((..---.......))))))..)))..)))))................------- ( -17.00, z-score = -3.87, R) >droPer1.super_36 714704 62 + 818889 CAGUGACUGCGUCGUCUGUUU---AAUUUUCACAGACAUCGCCUUCGCUUUCUCCCACACGCACA------- .(((((..(((..((((((..---.......))))))..)))..)))))................------- ( -17.00, z-score = -3.87, R) >consensus GAAUGGCUGCGUCGUCUGUUUUAUAAUUUUCACAGACGUCGCCCCCAUCACAAGCCCCCCCCCC________ ..((((..(((.(((((((............))))))).)))..))))........................ (-17.79 = -17.29 + -0.51)
| Location | 13,792,997 – 13,793,065 |
|---|---|
| Length | 68 |
| Sequences | 7 |
| Columns | 72 |
| Reading direction | reverse |
| Mean pairwise identity | 75.40 |
| Shannon entropy | 0.44939 |
| G+C content | 0.53070 |
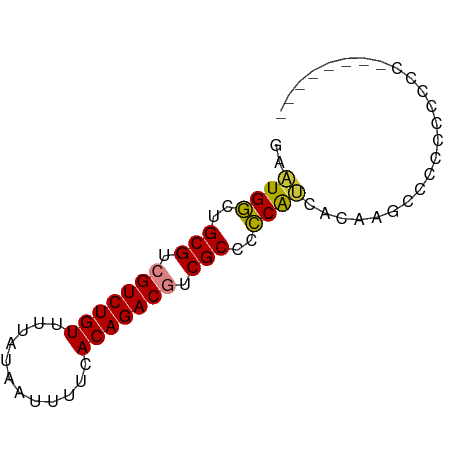
| Mean single sequence MFE | -21.01 |
| Consensus MFE | -20.55 |
| Energy contribution | -19.57 |
| Covariance contribution | -0.98 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.997915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13792997 68 - 24543557 ----GGUGGGGAGGGGGGCUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUC ----.(..((........))..)(((((..(((.(((((((............))))))).)))..))))). ( -21.70, z-score = -3.05, R) >droEre2.scaffold_4784 13799886 70 - 25762168 --GGGGAAAGUGGGGGGACUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUC --.......(..((....))..)(((((..(((.(((((((............))))))).)))..))))). ( -25.60, z-score = -4.95, R) >droYak2.chr3L 13893283 72 - 24197627 UGGGGGGAGGGGGGUGGACUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUC .......(.(((......))).)(((((..(((.(((((((............))))))).)))..))))). ( -21.30, z-score = -2.87, R) >droSim1.chr3L 13179058 62 - 22553184 ----------GGGGGGGUCUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUC ----------.............(((((..(((.(((((((............))))))).)))..))))). ( -20.70, z-score = -3.02, R) >droSec1.super_0 5954675 63 - 21120651 ---------AGGGGGGGUCUUGUGGUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUC ---------............(((((....(((.(((((((............))))))).))).))))).. ( -20.40, z-score = -2.63, R) >dp4.chrXR_group8 2643976 62 + 9212921 -------UGUGCGUGUGGGAGAAAGCGAAGGCGAUGUCUGUGAAAAUU---AAACAGACGACGCAGUCACUG -------.(((..((((.......((....))..(((((((.......---..))))))).))))..))).. ( -18.70, z-score = -2.14, R) >droPer1.super_36 714704 62 - 818889 -------UGUGCGUGUGGGAGAAAGCGAAGGCGAUGUCUGUGAAAAUU---AAACAGACGACGCAGUCACUG -------.(((..((((.......((....))..(((((((.......---..))))))).))))..))).. ( -18.70, z-score = -2.14, R) >consensus ________GGGGGGGGGGCUUGUGAUGGGGGCGACGUCUGUGAAAAUUAUAAAACAGACGACGCAGCCAUUC .......................(((((..(((.(((((((............))))))).)))..))))). (-20.55 = -19.57 + -0.98)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:28 2011