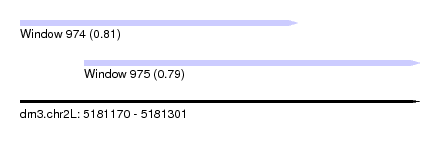
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,181,170 – 5,181,301 |
| Length | 131 |
| Max. P | 0.805793 |

| Location | 5,181,170 – 5,181,261 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 58.71 |
| Shannon entropy | 0.91224 |
| G+C content | 0.51643 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -4.56 |
| Energy contribution | -5.12 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.19 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.805793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
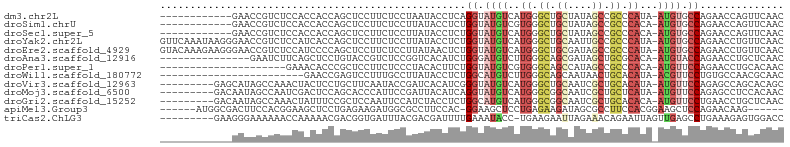
>dm3.chr2L 5181170 91 + 23011544 ------------GAACCGUCUCCACCACCAGCUCCUUCUCCUAAUACCUCAGGUAUGUCAUGGGCUGCUAUAGCCGCCCAUA-AUGUGCCAGAACCAGUUCAAC ------------((((.(..((.......((......))............(((((((.((((((.((....)).)))))).-))))))).))..).))))... ( -23.80, z-score = -2.75, R) >droSim1.chrU 4723875 91 - 15797150 ------------GAACCGUCUCCACCACCAGCUCCUUCUCCUUAUACCUCUGGUAUGUCGUGGGCUGCUAUAGCCGCCCACA-AUGUGCCAGAACCAGUUCAAC ------------((((.((....)).......................((((((((((.((((((.((....)).)))))).-))))))))))....))))... ( -30.00, z-score = -4.54, R) >droSec1.super_5 3263041 91 + 5866729 ------------GAACCGUCUCCACCACCAGCUCCUUCUCCUUAUACCUCUGGUAUGUCAUGGGCUGCUAUAGCCGCCCACA-AUGUGCCAGAACCAGUUCAAC ------------((((.((....)).......................((((((((((..(((((.((....)).)))))..-))))))))))....))))... ( -26.80, z-score = -3.70, R) >droYak2.chr2L 11978373 103 - 22324452 GUUCAAAUAAGGGAACCGUCUCCAUCACCAGCUCCUUCUCCUUAUACCUCUGGUAUGUCAUGGGCUGCAAUUGCCGCCCAUA-AUGUGCCAGAACCUGUUCAAC (..((.(((((((((....((........))....))).))))))...((((((((((.((((((.((....)).)))))).-))))))))))...))..)... ( -32.70, z-score = -4.13, R) >droEre2.scaffold_4929 5259385 103 + 26641161 GUACAAAGAAGGGAACCGUCUCCAUCCCCAGCUCCUUCUCCUUAUAACUCUGGUAUGUCAUGGGCUGCGAUAGCCGCCCAUA-AUGUGCCAGAACCUGUUCAAC (.(((.(((((((...................))))))).........((((((((((.((((((.((....)).)))))).-))))))))))...))).)... ( -33.61, z-score = -3.68, R) >droAna3.scaffold_12916 6197360 88 - 16180835 ---------------GAAUCUUCAGCUCCUGUACCGUCUCCGGUCACAUCUGGGAUGUCUUGGGCAGCGAUAGCUGCGCACA-AUGUACCAGAACCUGCUCAAC ---------------........(((...(((((((....)))).)))(((((.((((..((.(((((....))))).))..-)))).)))))....))).... ( -28.60, z-score = -1.85, R) >droPer1.super_1 6080235 82 - 10282868 ---------------------GAAACACCCGCUCCUUCUCCCUACACUUCUGGUAUGUCGUGGGCAGCCAUAGCCGCCCACA-AUGUUCCAGAACCUGCACAAC ---------------------..........................((((((.((((.((((((.((....)).)))))).-)))).)))))).......... ( -23.90, z-score = -3.23, R) >droWil1.scaffold_180772 7806749 79 - 8906247 ------------------------GAACCGAGUCCUUUGCCUUAUACCUCUGGCAUGUCUUGGGCAGCAAUAACUGCACAUA-ACGUUCCUGUGCCAACGCAAC ------------------------...(((((.(...((((..........)))).).)))))((((......)))).....-.......((((....)))).. ( -19.10, z-score = -1.01, R) >droVir3.scaffold_12963 13788504 94 - 20206255 ---------GAGCAUAGCCAAACUACUCCUGCUUCAAUACCGAUCACAUCGGGUAUGUCAUGGGCUGCAAUCGCUGCACAUA-AUGUUCCAGAGCCAGCACAGC ---------((((((((((..(((((((.((..((......))...))..))))).))....))))(((.....))).....-))))))............... ( -17.30, z-score = 1.79, R) >droMoj3.scaffold_6500 6788862 94 - 32352404 ---------GACAAUAGCCAAUCGACUCCAGCACCCAUUCCGAUUACAUCAGGUAUGUCAUGGGCGGCAAUCGCUGCUCAUA-AUGUUCCAGAGCCUCCACAAC ---------.......((((((((................)))))......((.((((.(((((((((....))))))))).-)))).)).).))......... ( -20.89, z-score = -0.77, R) >droGri2.scaffold_15252 7546506 94 - 17193109 ---------GACAAUAGCCAAACUAUUUCCGCUCCAAUUCCAUCUACCUCUGGCAUGUCAUGGGCGGCAAUCGCUGCACACA-AUGUUCCUGAACCUGCUCAAC ---------((((...((((..............................)))).)))).(((((((...(((..(((....-.)))...)))..))))))).. ( -16.71, z-score = 0.11, R) >apiMel3.Group3 4332782 91 + 12341916 ------AUGGCGACUUCCACGGAAGCUCCUGAGAAGAUGGCGCCUUCCAC-GGAAGCUCCUGAGAAGAUAGCGCCUUCCACGGAAGCUCCAGAACAAG------ ------.(((.(.(((((..((.....))......((.(((((((((((.-((.....)))).))))...))))).))...)))))).))).......------ ( -29.70, z-score = -1.19, R) >triCas2.ChLG3 29406274 94 + 32080666 ---------GAAGGGAAAAAACCAAAAACGACGGUGAUUUACGACGAUUUUGAAAUACC-UGAAGAAUUAGAAACAGAAUUAGUUGAGCCUGAAAGAGUGGACC ---------............(((........(((......(((((((((((......(-(((....))))...))))))).)))).)))........)))... ( -10.89, z-score = 0.17, R) >consensus ____________GAACCGUCUCCAACACCAGCUCCUUCUCCUUAUACCUCUGGUAUGUCAUGGGCUGCAAUAGCCGCCCAUA_AUGUGCCAGAACCAGCUCAAC ...................................................((.((((..((.((.((....)).)).))...)))).)).............. ( -4.56 = -5.12 + 0.56)


| Location | 5,181,191 – 5,181,301 |
|---|---|
| Length | 110 |
| Sequences | 13 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 61.97 |
| Shannon entropy | 0.86772 |
| G+C content | 0.46923 |
| Mean single sequence MFE | -26.71 |
| Consensus MFE | -5.55 |
| Energy contribution | -5.67 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
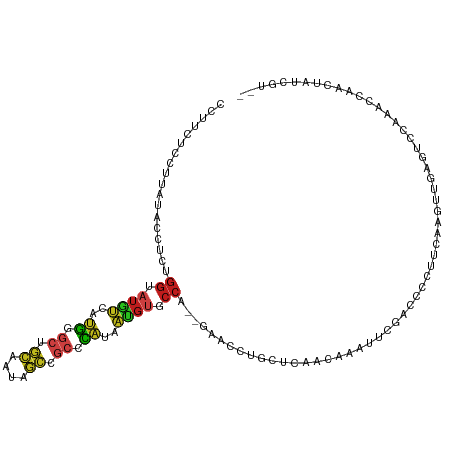
>dm3.chr2L 5181191 110 + 23011544 CCUUCUCCUAAUACCUCAGGUAUGUCAUGGGCUGCUAUAGCCGCCCAUAAUGUGCCA--GAACCAGUUCAACAUAUUCGACCCCUCCAAGCUGAGGCCAAACCAACUGUCGU-- .............((((((((((((.((((((.((....)).)))))).))))))).--(((....)))......................)))))................-- ( -29.50, z-score = -2.40, R) >droSim1.chrU 4723896 109 - 15797150 CCUUCUCCUUAUACCUCUGGUAUGUCGUGGGCUGCUAUAGCCGCCCACAAUGUGCCA--GAACCAGUUCAACAUAUUCGACCCCUGCA-GUUGAGUCCAAACAAACUUUCCU-- ...............((((((((((.((((((.((....)).)))))).))))))))--))...((((......(((((((.......-))))))).......)))).....-- ( -33.42, z-score = -4.54, R) >droSec1.super_5 3263062 110 + 5866729 CCUUCUCCUUAUACCUCUGGUAUGUCAUGGGCUGCUAUAGCCGCCCACAAUGUGCCA--GAACCAGUUCAACAUAUUCGACCCCUUCAAGUUGAGUCCAAACCAACUGUCGU-- ...............((((((((((..(((((.((....)).)))))..))))))))--))..(((((......(((((((........))))))).......)))))....-- ( -33.62, z-score = -4.63, R) >droYak2.chr2L 11978406 110 - 22324452 CCUUCUCCUUAUACCUCUGGUAUGUCAUGGGCUGCAAUUGCCGCCCAUAAUGUGCCA--GAACCUGUUCAACAAAUUCGACCCCUUCCAGUUGAGGCCAAGCCAACUAUCGU-- ...............((((((((((.((((((.((....)).)))))).))))))))--))...............(((((........)))))(((...))).........-- ( -33.10, z-score = -3.69, R) >droEre2.scaffold_4929 5259418 110 + 26641161 CCUUCUCCUUAUAACUCUGGUAUGUCAUGGGCUGCGAUAGCCGCCCAUAAUGUGCCA--GAACCUGUUCAACAUAUUCGACCCCUUCAAGUUGAGUCCAAACCAACUAUCGU-- ...............((((((((((.((((((.((....)).)))))).))))))))--)).............(((((((........)))))))................-- ( -31.90, z-score = -4.30, R) >droAna3.scaffold_12916 6197378 110 - 16180835 CCGUCUCCGGUCACAUCUGGGAUGUCUUGGGCAGCGAUAGCUGCGCACAAUGUACCA--GAACCUGCUCAACAAAUAAGACCCCUUUUGGUAGAAACGAAAUCGGUUGUGGU-- (((....)))((((.(((((.((((..((.(((((....))))).))..)))).)))--)).................((((..(((((.......)))))..)))))))).-- ( -32.60, z-score = -0.76, R) >droPer1.super_1 6080247 110 - 10282868 CCUUCUCCCUACACUUCUGGUAUGUCGUGGGCAGCCAUAGCCGCCCACAAUGUUCCA--GAACCUGCACAACAAAUACGUCCGCUUCAAAUUGAAACCAAGUCCAUAAUUGU-- ..............((((((.((((.((((((.((....)).)))))).)))).)))--)))........((((.((.(..(..(((.....))).....)..).)).))))-- ( -24.60, z-score = -1.97, R) >droWil1.scaffold_180772 7806758 110 - 8906247 CCUUUGCCUUAUACCUCUGGCAUGUCUUGGGCAGCAAUAACUGCACAUAACGUUCCU--GUGCCAACGCAACAAAUACGUCCUCUAUUGAUCGCGACUAAAGAGAUUGUAAA-- ....((((..........))))(((.((((((((......)))).((((.......)--))))))).))).....((((..((((.(((........)))))))..))))..-- ( -21.60, z-score = -0.08, R) >droVir3.scaffold_12963 13788528 110 - 20206255 UCAAUACCGAUCACAUCGGGUAUGUCAUGGGCUGCAAUCGCUGCACAUAAUGUUCCA--GAGCCAGCACAGCAAAUUCGUCUCCUUCAAAUUGAAUCUACA--GAUAAAAUUGU (((((...((......(((((.(((..((.((((...((...(((.....)))....--))..)))).))))).)))))......))..)))))(((....--)))........ ( -16.50, z-score = 1.69, R) >droMoj3.scaffold_6500 6788886 110 - 32352404 CCCAUUCCGAUUACAUCAGGUAUGUCAUGGGCGGCAAUCGCUGCUCAUAAUGUUCCA--GAGCCUCCACAACAAAUCAGACUUAUUCAAAUUGAAUCAAAACCAUCAACAUU-- ........((((.((...((((.((((((((((((....)))))))))..((((...--(.....)...)))).....))).)))).....))))))...............-- ( -20.20, z-score = -0.87, R) >droGri2.scaffold_15252 7546530 110 - 17193109 CCAAUUCCAUCUACCUCUGGCAUGUCAUGGGCGGCAAUCGCUGCACACAAUGUUCCU--GAACCUGCUCAACAAAUACGACCUCUUCAAAUUGAAACUAAGGAAAUUACUUU-- ....((((.........(((((..(((.(((((((....)))))((.....)).)))--))...))).))..............(((.....))).....))))........-- ( -18.40, z-score = -0.25, R) >apiMel3.Group3 4332803 107 + 12341916 -----CCUGAGAAGAUGGCGCCUUCCACGGAAGCUCCUGAGAAGAUAGCGCCUUCCACGGAAGCUCCAGAACAAGAAAUUUCUUCCGAGGCUCCGUCAGAGAAAACUGCAAC-- -----.((((...((.(((((((((((.((.....)))).))))...))))).))..((((.((..(.(((.((....))..))).)..)))))))))).............-- ( -31.00, z-score = -0.81, R) >triCas2.ChLG3 29406298 105 + 32080666 GUGAUUUACGACGAUUUUGAAAUACCUGAAGAAUUAGAAACAGAAUUAGUUGAGCCU--GAAAGAGUGGACCACAUUGUUGAGUUACCUGCUGAAAUCCAGGUAACU------- (((.....(((((((((((......((((....))))...))))))).))))..((.--........))..))).......(((((((((........)))))))))------- ( -20.80, z-score = -1.66, R) >consensus CCUUCUCCUUAUACCUCUGGUAUGUCAUGGGCUGCAAUAGCCGCCCAUAAUGUGCCA__GAACCUGCUCAACAAAUUCGACCCCUUCAAGUUGAGUCCAAACCAACUAUCGU__ ..................((.((((..((.((.((....)).)).))..)))).)).......................................................... ( -5.55 = -5.67 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:16 2011