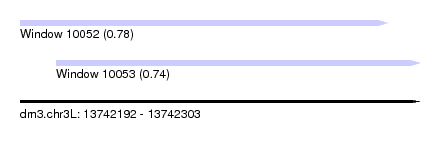
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,742,192 – 13,742,303 |
| Length | 111 |
| Max. P | 0.782750 |

| Location | 13,742,192 – 13,742,294 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.36 |
| Shannon entropy | 0.39729 |
| G+C content | 0.42056 |
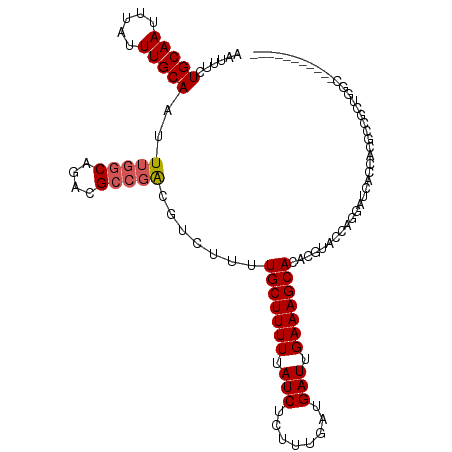
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -16.11 |
| Energy contribution | -16.46 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
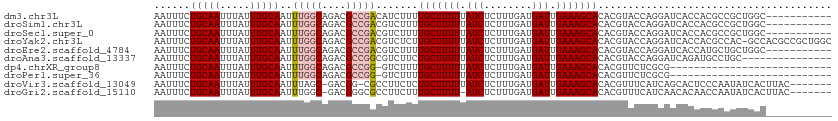
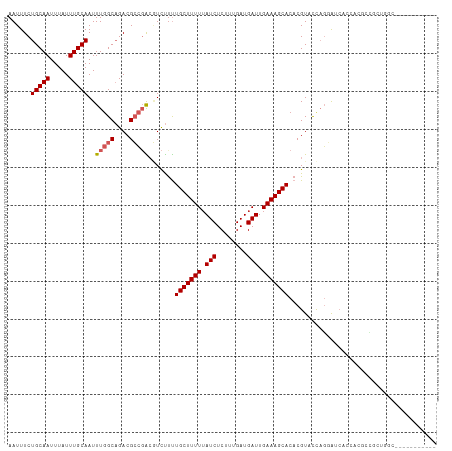
>dm3.chr3L 13742192 102 + 24543557 AAUUUCUGCAAUUUAUUUGCAAUUUGGCAGACGCCGACAUCUUUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUACCAGGAUCACCACGCCGCUGGC----------- ......(((((.....)))))..(((((....))))).......(((((((.(((........))).)))))))......((((.............)))).----------- ( -22.02, z-score = -0.88, R) >droSim1.chr3L 13127659 102 + 22553184 AAUUUCUGCAAUUUAUUUGCAAUUUGGCAGACGCCGACGUCUUUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUACCAGGAUCACCACGCCGCUGGC----------- ......(((((.....)))))....((((((((....)))))..(((((((.(((........))).))))))).........((....))..)))......----------- ( -24.10, z-score = -1.21, R) >droSec1.super_0 5903967 102 + 21120651 AAUUUCUGCAAUUUAUUUGCAAUUUGGCAGACGCCGACGUCUUUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUACCAGGAUCACCACGCCGCUGGC----------- ......(((((.....)))))....((((((((....)))))..(((((((.(((........))).))))))).........((....))..)))......----------- ( -24.10, z-score = -1.21, R) >droYak2.chr3L 13839870 112 + 24197627 AAUUUCUGCAAUUUAUUUGCAAUUUGGCAGACGCCGACGUCUCUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUACCAGGAUCACCACGCCAC-GCCACGCCGCUGGC ......(((((.....)))))...(((((((((....)))))..(((((((.(((........))).))))))).........((....))..)))).-((((......)))) ( -28.60, z-score = -1.65, R) >droEre2.scaffold_4784 13748683 102 + 25762168 AAUUUCUGCAAUUUAUUUGCAAUUUGGCAGACGCCGACGUCUUUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUACCAGGAUCACCAUGCUGCUGGC----------- ......(((((.....)))))...((((....))))((((....(((((((.(((........))).))))))).)))).((((.(.(.....).).)))).----------- ( -25.00, z-score = -1.41, R) >droAna3.scaffold_13337 1619778 98 - 23293914 AAUUUCUGCAAUUUAUUUGCAAUUUGGCAGACGCCGGCGUCUUCUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUACCAGGAUCAGAUGCCUGC--------------- ......(((((.....)))))....(((....)))(((((((..(((((((.(((........))).)))))))..(......)....)))))))...--------------- ( -27.10, z-score = -1.91, R) >dp4.chrXR_group8 2534627 85 - 9212921 AAUUUCUGCAAUUUAUUUGCAAUUUGGCAGACGCCGG-GUCUUUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUUCUCGCG--------------------------- ......(((((.....)))))....(((....)))((-(.....(((((((.(((........))).)))))))......)))...--------------------------- ( -20.10, z-score = -1.57, R) >droPer1.super_36 630258 85 + 818889 AAUUUCUGCAAUUUAUUUGCAAUUUGGCAGACGCCGG-GUCUUUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUUCUCGCG--------------------------- ......(((((.....)))))....(((....)))((-(.....(((((((.(((........))).)))))))......)))...--------------------------- ( -20.10, z-score = -1.57, R) >droVir3.scaffold_13049 5590366 104 - 25233164 AAUUUCUGCAAUUUAUUUGCAAUUUAGC-GACGG-CGCCUUCUCUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUUUCAUCAGCACUCCCAAUAUCACUUAC------- ......(((((.....))))).....((-((.((-.((......(((((((.(((........))).)))))))...)).)).)).))..................------- ( -16.20, z-score = -0.67, R) >droGri2.scaffold_15110 17712995 104 + 24565398 AAUUUCUGCAAUUUAUUUGCAAUUUGGC-GACGGGCGCCUUCUUUGCUUUU-AUCUCUUUGAUGAUUGAAAGCACACGUUUCAUCAACACAACCAAUAUCACUUAC------- ......(((((.....)))))....(((-(.....)))).....(((((((-(((........)).))))))))................................------- ( -18.40, z-score = -1.40, R) >consensus AAUUUCUGCAAUUUAUUUGCAAUUUGGCAGACGCCGACGUCUUUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUACCAGGAUCACCACGCCGCUGGC___________ ......(((((.....)))))..(((((....))))).......(((((((.(((........))).)))))))....................................... (-16.11 = -16.46 + 0.35)
| Location | 13,742,202 – 13,742,303 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.52 |
| Shannon entropy | 0.48352 |
| G+C content | 0.43705 |
| Mean single sequence MFE | -21.01 |
| Consensus MFE | -12.05 |
| Energy contribution | -12.25 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13742202 101 + 24543557 AUUUAUUUGCAAUUUGGCAGACGCCGACAUCUUUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUACCAGGAUCACCACGCC----------GCUGGCCUUCCUUCA- .......(((...(((((....))))).......(((((((.(((........))).)))))))...)))..((((...(((....----------..)))...))))...- ( -21.60, z-score = -1.29, R) >droSim1.chr3L 13127669 101 + 22553184 AUUUAUUUGCAAUUUGGCAGACGCCGACGUCUUUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUACCAGGAUCACCACGCC----------GCUGGCCUUCCUUCA- ..............((((....))))((((....(((((((.(((........))).))))))).))))...((((...(((....----------..)))...))))...- ( -22.00, z-score = -1.02, R) >droSec1.super_0 5903977 101 + 21120651 AUUUAUUUGCAAUUUGGCAGACGCCGACGUCUUUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUACCAGGAUCACCACGCC----------GCUGGCCUUCCUUCA- ..............((((....))))((((....(((((((.(((........))).))))))).))))...((((...(((....----------..)))...))))...- ( -22.00, z-score = -1.02, R) >droYak2.chr3L 13839880 112 + 24197627 AUUUAUUUGCAAUUUGGCAGACGCCGACGUCUCUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUACCAGGAUCACCACGCCACGCCACGCCGCUGGCCUUCCUCCUA ..............((((....))))((((....(((((((.(((........))).))))))).))))...((((.......((((.((......)))))).....)))). ( -26.00, z-score = -1.39, R) >droEre2.scaffold_4784 13748693 101 + 25762168 AUUUAUUUGCAAUUUGGCAGACGCCGACGUCUUUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUACCAGGAUCACCAUGCU----------GCUGGCCUUCCUCCA- ..............((((....))))((((....(((((((.(((........))).))))))).))))...((((...(((....----------..)))...))))...- ( -22.10, z-score = -0.91, R) >droAna3.scaffold_13337 1619788 97 - 23293914 AUUUAUUUGCAAUUUGGCAGACGCCGGCGUCUUCUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUACCAGGAUCA-GAUGCCUG-------CACUCCCCUU------- .......((((....(((....)))(((((((..(((((((.(((........))).)))))))..(......)....)-))))))))-------))........------- ( -26.30, z-score = -2.23, R) >dp4.chrXR_group8 2534637 80 - 9212921 AUUUAUUUGCAAUUUGGCAGACGCCGG-GUCUUUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUUCUCGCGCUCUG------------------------------- .....(((((......))))).(((((-(.....(((((((.(((........))).)))))))......)))).))....------------------------------- ( -19.90, z-score = -1.75, R) >droPer1.super_36 630268 80 + 818889 AUUUAUUUGCAAUUUGGCAGACGCCGG-GUCUUUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUUCUCGCGCUCUA------------------------------- .....(((((......))))).(((((-(.....(((((((.(((........))).)))))))......)))).))....------------------------------- ( -19.90, z-score = -1.84, R) >droVir3.scaffold_13049 5590376 94 - 25233164 AUUUAUUUGCAAUUUAGC-GACGG-CGCCUUCUCUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUUUCAUCAGCACUCCCAAUA------UCACUUAC---------- .......(((......((-(....-)))......(((((((.(((........))).)))))))............))).........------........---------- ( -14.50, z-score = -0.84, R) >droGri2.scaffold_15110 17713005 99 + 24565398 AUUUAUUUGCAAUUUGGC-GACGGGCGCCUUCUUUGCUUUU-AUCUCUUUGAUGAUUGAAAGCACACGUUUCAUCAACACAACCAAUA------UCACUUACCAACA----- ........((((...(((-(.....))))....))))....-......(((((((.((........))..)))))))...........------.............----- ( -15.80, z-score = -0.99, R) >consensus AUUUAUUUGCAAUUUGGCAGACGCCGACGUCUUUUGCUUUUUAUCUCUUUGAUGAUUGAAAGCACACGUACCAGGAUCACCACGCC__________GCUGGCCUUCC_____ .....(((((......))))).............(((((((.(((........))).)))))))................................................ (-12.05 = -12.25 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:20 2011