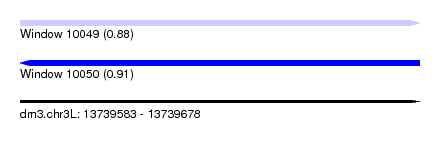
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,739,583 – 13,739,678 |
| Length | 95 |
| Max. P | 0.909329 |

| Location | 13,739,583 – 13,739,678 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 91.28 |
| Shannon entropy | 0.16863 |
| G+C content | 0.40441 |
| Mean single sequence MFE | -18.67 |
| Consensus MFE | -17.06 |
| Energy contribution | -17.11 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13739583 95 + 24543557 ---UAUCUCGACAUCAGCUGUUUAUUCUUGUUUGUCGCUGAUGUCGCUUCAUUAAACGGUAAAGUUAUUAAAUUACCACAACCAUCGAACC-GACCAAC- ---.....((((((((((..(..((....))..)..))))))))))...........(((((..........)))))..............-.......- ( -19.00, z-score = -1.97, R) >droPer1.super_36 627319 100 + 818889 CCUUCUCCCGGCAUCAGCUGUUUAUUCUUGUUUGUUGCUGAUGUCGCUUCAUUAAAUGGCAAAGUUAUUAAAUUACCGCAACCAUCGAACCGACCGAACA .........(((((((((..(..((....))..)..)))))))))((...(((.((((((...)))))).)))....)).....(((.......)))... ( -17.70, z-score = -0.30, R) >dp4.chrXR_group8 2531661 100 - 9212921 CCUUCUCCCGACAUCAGCUGUUUAUUCUUGUUUGUUGCUGAUGUCGCUUCAUUAAAUGGCAAAGUUAUUAAAUUACCGCAACCAUCGAACCGACCGAACA .........(((((((((..(..((....))..)..)))))))))((...(((.((((((...)))))).)))....)).....(((.......)))... ( -18.30, z-score = -1.12, R) >droAna3.scaffold_13337 1617706 95 - 23293914 ---UUUGUCGACAUCAGCUGUUUAUUCUUAUUUGCCGCUGAUGUCGCUUCAUUAAAUGGUAGAGUUAUUAAAUUACCGCAGCCAUCGAACC-AGCCAAC- ---......(((((((((.((..((....))..)).)))))))))(((...((..(((((.(.(.((......)).).).)))))..))..-)))....- ( -23.50, z-score = -2.51, R) >droEre2.scaffold_4784 13746098 96 + 25762168 ---UCUCUCGACAUCAGCUGUUUAUUCUUGUUUGUCGCUGAUGUCGCUUCAUUAAACGGUAAAGUUAUUAAAUUACCACAACCAUCGAACCCAACCAAC- ---.....((((((((((..(..((....))..)..))))))))))...........(((((..........)))))......................- ( -19.00, z-score = -2.62, R) >droYak2.chr3L 13837304 95 + 24197627 ---UCUCUCGACAUCAGCUGUUUAUUCUUGUUUGUCGCUGAUGUCGCUUCAUUAAACGGUAAAGUUAUUAAAUUACCACAACCAUCGAACC-GACCAAC- ---.....((((((((((..(..((....))..)..))))))))))...........(((((..........)))))..............-.......- ( -19.00, z-score = -1.99, R) >droSec1.super_0 5901389 95 + 21120651 ---UCUCUCAACAUCAGCUGUUUAUUCUUGUUUGUCGCUGAUGUCGCUUCAUUAAACGGUAAAGUUAUUAAAUUACCACAACCAUCGAACC-GACCAAC- ---.........((((((..(..((....))..)..))))))((((.(((.......(((((..........))))).........))).)-)))....- ( -13.89, z-score = -0.62, R) >droSim1.chr3L 13125078 95 + 22553184 ---UCUCUCGACAUCAGCUGUUUAUUCUUGUUUGUCGCUGAUGUCGCUUCAUUAAACGGUAAAGUUAUUAAAUUACCACAACCAUCGAACC-GACCAAC- ---.....((((((((((..(..((....))..)..))))))))))...........(((((..........)))))..............-.......- ( -19.00, z-score = -1.99, R) >consensus ___UCUCUCGACAUCAGCUGUUUAUUCUUGUUUGUCGCUGAUGUCGCUUCAUUAAACGGUAAAGUUAUUAAAUUACCACAACCAUCGAACC_AACCAAC_ ........((((((((((..(..((....))..)..))))))))))...........(((((..........)))))....................... (-17.06 = -17.11 + 0.05)

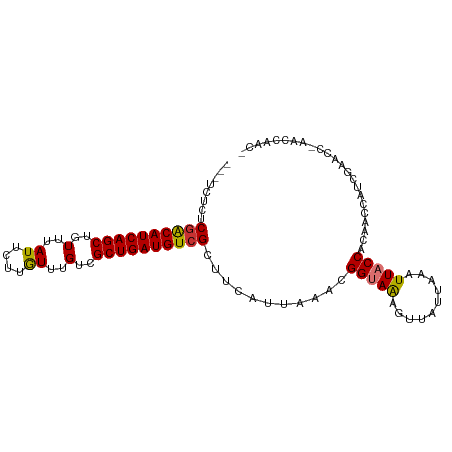
| Location | 13,739,583 – 13,739,678 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 91.28 |
| Shannon entropy | 0.16863 |
| G+C content | 0.40441 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.17 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.909329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13739583 95 - 24543557 -GUUGGUC-GGUUCGAUGGUUGUGGUAAUUUAAUAACUUUACCGUUUAAUGAAGCGACAUCAGCGACAAACAAGAAUAAACAGCUGAUGUCGAGAUA--- -(((((.(-(((.....((((((.........))))))..)))).)))))....((((((((((..................)))))))))).....--- ( -26.27, z-score = -2.39, R) >droPer1.super_36 627319 100 - 818889 UGUUCGGUCGGUUCGAUGGUUGCGGUAAUUUAAUAACUUUGCCAUUUAAUGAAGCGACAUCAGCAACAAACAAGAAUAAACAGCUGAUGCCGGGAGAAGG ..((((((((.(((((((((...(((.........)))..))))))....))).)))(((((((..................))))))))))))...... ( -23.77, z-score = -0.82, R) >dp4.chrXR_group8 2531661 100 + 9212921 UGUUCGGUCGGUUCGAUGGUUGCGGUAAUUUAAUAACUUUGCCAUUUAAUGAAGCGACAUCAGCAACAAACAAGAAUAAACAGCUGAUGUCGGGAGAAGG ..(((..(((.(((((((((...(((.........)))..))))))....))).)(((((((((..................)))))))))))..))).. ( -24.47, z-score = -1.30, R) >droAna3.scaffold_13337 1617706 95 + 23293914 -GUUGGCU-GGUUCGAUGGCUGCGGUAAUUUAAUAACUCUACCAUUUAAUGAAGCGACAUCAGCGGCAAAUAAGAAUAAACAGCUGAUGUCGACAAA--- -(((.(((-.(((.(((((.((.(((.........))).))))))).)))..)))(((((((((..................))))))))))))...--- ( -25.37, z-score = -2.07, R) >droEre2.scaffold_4784 13746098 96 - 25762168 -GUUGGUUGGGUUCGAUGGUUGUGGUAAUUUAAUAACUUUACCGUUUAAUGAAGCGACAUCAGCGACAAACAAGAAUAAACAGCUGAUGUCGAGAGA--- -....(((..(((.((((((...(((.........)))..)))))).)))..)))(((((((((..................)))))))))......--- ( -24.87, z-score = -1.88, R) >droYak2.chr3L 13837304 95 - 24197627 -GUUGGUC-GGUUCGAUGGUUGUGGUAAUUUAAUAACUUUACCGUUUAAUGAAGCGACAUCAGCGACAAACAAGAAUAAACAGCUGAUGUCGAGAGA--- -(((((.(-(((.....((((((.........))))))..)))).)))))....((((((((((..................)))))))))).....--- ( -26.27, z-score = -2.26, R) >droSec1.super_0 5901389 95 - 21120651 -GUUGGUC-GGUUCGAUGGUUGUGGUAAUUUAAUAACUUUACCGUUUAAUGAAGCGACAUCAGCGACAAACAAGAAUAAACAGCUGAUGUUGAGAGA--- -(((((.(-(((.....((((((.........))))))..)))).)))))....((((((((((..................)))))))))).....--- ( -24.17, z-score = -1.80, R) >droSim1.chr3L 13125078 95 - 22553184 -GUUGGUC-GGUUCGAUGGUUGUGGUAAUUUAAUAACUUUACCGUUUAAUGAAGCGACAUCAGCGACAAACAAGAAUAAACAGCUGAUGUCGAGAGA--- -(((((.(-(((.....((((((.........))))))..)))).)))))....((((((((((..................)))))))))).....--- ( -26.27, z-score = -2.26, R) >consensus _GUUGGUC_GGUUCGAUGGUUGUGGUAAUUUAAUAACUUUACCGUUUAAUGAAGCGACAUCAGCGACAAACAAGAAUAAACAGCUGAUGUCGAGAGA___ ..........(((.((((((...(((.........)))..)))))).)))....((((((((((..................))))))))))........ (-22.42 = -22.17 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:17 2011