
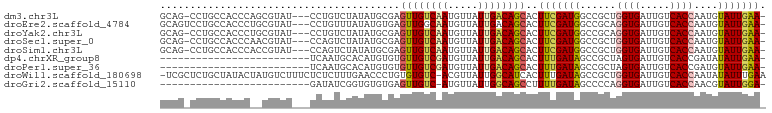
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,729,229 – 13,729,325 |
| Length | 96 |
| Max. P | 0.807287 |

| Location | 13,729,229 – 13,729,325 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 72.56 |
| Shannon entropy | 0.53195 |
| G+C content | 0.45656 |
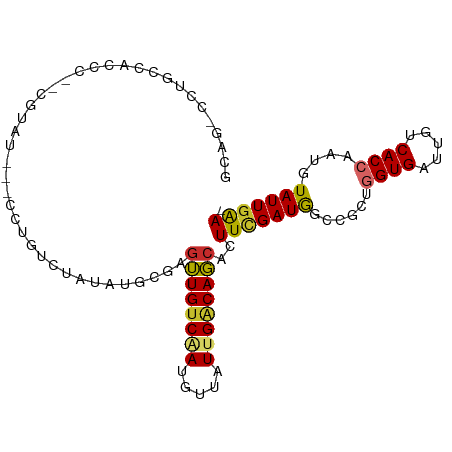
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -13.48 |
| Energy contribution | -13.19 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.807287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13729229 96 - 24543557 GCAG-CCUGCCACCCAGCGUAU---CCUGUCUAUAUGCGAGUUGUCAAUGUUAUUGACAGCACUUCGAUGGCCGCUGGUGAUUGUCACCAAUGUAUUGAA- ((((-(..((((....((((((---.......))))))..(((((((((...))))))))).......)))).))(((((.....))))).)))......- ( -28.70, z-score = -1.50, R) >droEre2.scaffold_4784 13735689 97 - 25762168 GCAGUCCUGCCACCCUGCGUAU---CCUGUUUAUAUGUGAGUUGGCAAUGUUAUUGACAGCACUUCGAUGGCCGCAGGUGAUUGUCACCAAUGUAUUGAA- .((((..(((((.(..((((((---.......))))))..).)))))(((((..(((((((((((((.....)).))))).))))))..)))))))))..- ( -25.60, z-score = -0.26, R) >droYak2.chr3L 13826582 96 - 24197627 GCAG-CCUGCCACCCUGCGUAU---CCUGUCUAUAUGCGAGUUGUCAAUGUUAUUGACAGCACUUCGAUGGCCGCAGGUGAUUGUCACCAAUGUAUUGAA- ((((-(..((((...(((((((---.......))))))).(((((((((...))))))))).......)))).)).((((.....))))..)))......- ( -28.50, z-score = -1.43, R) >droSec1.super_0 5891054 96 - 21120651 GCAG-CCUGCCACCCAACGUAU---CCAGUCUAUAUGCGAGUUGUCAAUGUUAUUGACAGCACUUCGAUGGCCGCUGGUGAUUGUCACCAAUGUAUUGAA- ((((-(..((((.....(((((---.........))))).(((((((((...))))))))).......)))).))(((((.....))))).)))......- ( -26.30, z-score = -1.02, R) >droSim1.chr3L 13114867 96 - 22553184 GCAG-CCUGCCACCCACCGUAU---CCAGUCUAUAUGCGAGUUGUCAAUGUUAUUGACAGCACUUCGAUGGCCGCUGGUGAUUGUCACCAAUGUAUUGAA- ((((-(..((((.....(((((---.........))))).(((((((((...))))))))).......)))).))(((((.....))))).)))......- ( -26.10, z-score = -0.97, R) >dp4.chrXR_group8 2520961 75 + 9212921 -------------------------UCAAUGCACAUGUGUGUUGUCGAUGUUAUUGACAGCACUUUGAUAGCCGCUAGUGAUUGUCACCGAUAUAUUGAA- -------------------------((((((.......(((((((((((...)))))))))))..(((((..((....))..)))))......)))))).- ( -20.40, z-score = -1.45, R) >droPer1.super_36 616640 75 - 818889 -------------------------UCAAUGCACAUGUGUGUUGUCGAUGUUAUUGACAGCACUUUGAUAGCCGCUAGUGAUUGUCACCGAUGUAUUGAA- -------------------------(((((((((....(((((((((((...)))))))))))..(((((..((....))..)))))..).)))))))).- ( -25.10, z-score = -2.87, R) >droWil1.scaffold_180698 439296 99 - 11422946 -UCGCUCUGCUAUACUAUGUCUUUCUCUCUUUGAACCCUGUGUGUC-ACGUUAUUGGCAUCACUUUGAUAGCCGCUGGUGAUUGUCACCAAUAUAUUUGAA -..((...(((((.........(((.......)))....(((((((-(......))))).)))....))))).))(((((.....)))))........... ( -19.20, z-score = -1.03, R) >droGri2.scaffold_15110 17702525 74 - 24565398 -------------------------GAUAUCGGUGUGUGAGUUGUC-AUGUUAUUGGCAGCCUUUUGAUAGCCCCAGGUGAUUGUCACCAACGUAUUGGA- -------------------------....(((((((((..((((((-(......)))))))...............((((.....)))).))))))))).- ( -18.60, z-score = -0.47, R) >consensus GCAG_CCUGCCACCC__CGUAU___CCUGUCUAUAUGCGAGUUGUCAAUGUUAUUGACAGCACUUCGAUGGCCGCUGGUGAUUGUCACCAAUGUAUUGAA_ ........................................((((((((.....))))))))..(((((((......((((.....))))....))))))). (-13.48 = -13.19 + -0.29)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:16 2011