
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,721,983 – 13,722,110 |
| Length | 127 |
| Max. P | 0.745127 |

| Location | 13,721,983 – 13,722,110 |
|---|---|
| Length | 127 |
| Sequences | 9 |
| Columns | 127 |
| Reading direction | forward |
| Mean pairwise identity | 91.94 |
| Shannon entropy | 0.16733 |
| G+C content | 0.46297 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -24.31 |
| Energy contribution | -24.98 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
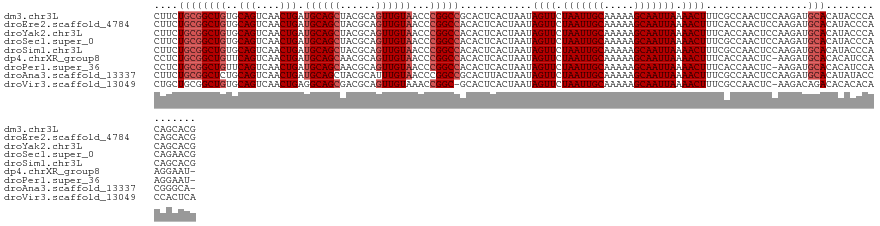
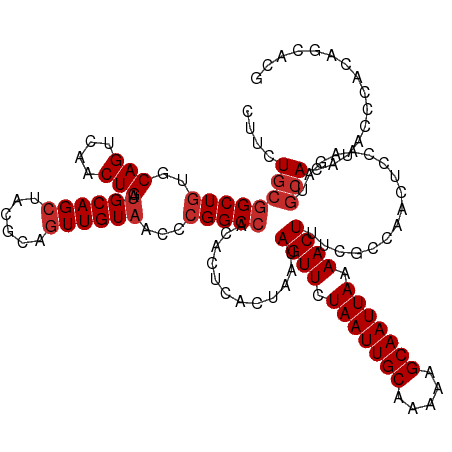
>dm3.chr3L 13721983 127 + 24543557 CUUCUGCGGCUGUGCAGUCAACUGAUGCAGCUACGCAGUUGUAACCCGGCCGCACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCGCCAACUCCAAGAUGCACAUACCCACAGCACG ..((((((((((..(((....))).(((((((....)))))))...)))))))..........((((.(((((((.....))))))).)))).............)))(((...........))).. ( -32.20, z-score = -1.73, R) >droEre2.scaffold_4784 13728321 127 + 25762168 CUUCUGCGGCUGUGCAGUCAACUGAUGCAGCUACGCAGUUGUAACCCGGCCACACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCACCAACUCCAAGAUGCACAUACCCACAGCACG ...(((.((.((((((.((..(((.(((((((....)))))))...)))..............((((.(((((((.....))))))).))))..............))))))))...)).))).... ( -29.30, z-score = -1.68, R) >droYak2.chr3L 13819095 127 + 24197627 CUUCUGCGGCUGUGCAGUCAACUGAUGCAGCUACGCAGUUGUAACCCGGCCACACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCACCAACUCCAAGAUGCACAUACCCACAGCACG ...(((.((.((((((.((..(((.(((((((....)))))))...)))..............((((.(((((((.....))))))).))))..............))))))))...)).))).... ( -29.30, z-score = -1.68, R) >droSec1.super_0 5884017 127 + 21120651 CUUCUGCGGCUGUGCAGUCAACUGAUGCAGCUACGCAGUUGUAACCCGGCCACACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCGCCAACUCCAAGAUGCACAUACCCACAGAACG .(((((.((.((((((.((......(((((((....)))))))....(((.............((((.(((((((.....))))))).))))...)))........))))))))...)).))))).. ( -34.69, z-score = -3.27, R) >droSim1.chr3L 13107726 127 + 22553184 CUUCUGCGGCUGUGCAGUCAACUGAUGCAGCUACGCAGUUGUAACCCGGCCACACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCGCCAACUCCAAGAUGCACAUACCCACAGCACG ...(((.((.((((((.((......(((((((....)))))))....(((.............((((.(((((((.....))))))).))))...)))........))))))))...)).))).... ( -31.79, z-score = -2.06, R) >dp4.chrXR_group8 2512714 125 - 9212921 CCUCUGCGGCUGUUCAGUCAACUGAUGCAGCAACGCAGUUGUAACCCGGCCACACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCACCAACUC-AAGAUGCACACAUCCAAGGAAU- ...((((((((((((((....)))).)))))..))))).......((................((((.(((((((.....))))))).))))...........-..((((....))))...))...- ( -30.70, z-score = -2.59, R) >droPer1.super_36 608422 125 + 818889 CCUCUGCGGCUGUUCAGUCAACUGAUGCAGCAACGCAGUUGUAACCCGGCCACACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCACCAACUC-AAGAUGCACACAUCCAAGGAAU- ...((((((((((((((....)))).)))))..))))).......((................((((.(((((((.....))))))).))))...........-..((((....))))...))...- ( -30.70, z-score = -2.59, R) >droAna3.scaffold_13337 1594959 126 - 23293914 CUUCUGCGGCUCUGCAGUCAACUGAUGCAGCUACGCAUUUGUAACCCGGCCGCACUUACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCGCCAACUCCAAGAUGCACAUAUACCCGGGCA- ....((((...(((((.((....)))))))...)))).......(((((..............((((.(((((((.....))))))).))))...((............))........)))))..- ( -30.50, z-score = -1.23, R) >droVir3.scaffold_13049 5568648 125 - 25233164 CUGCUGCGGCUGUGCAGUCAACUGAGGCAGCGACGCAGUUGUAAACCGGC-GCACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCGCCAACUC-AAGACAGACACACACACCACUCA (((((((((((((.(((....)))..)))))..))))).........(((-(...........((((.(((((((.....))))))).))))..)))).....-....)))................ ( -33.02, z-score = -1.31, R) >consensus CUUCUGCGGCUGUGCAGUCAACUGAUGCAGCUACGCAGUUGUAACCCGGCCACACUCACUAAUAGUUCUAAUUGCAAAAAGCAAUUAAAACUUUCGCCAACUCCAAGAUGCACAUACCCACAGCACG ....((((((((..(((....))).((((((......))))))...)))))............((((.(((((((.....))))))).)))).................)))............... (-24.31 = -24.98 + 0.67)
| Location | 13,721,983 – 13,722,110 |
|---|---|
| Length | 127 |
| Sequences | 9 |
| Columns | 127 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Shannon entropy | 0.16733 |
| G+C content | 0.46297 |
| Mean single sequence MFE | -44.04 |
| Consensus MFE | -31.58 |
| Energy contribution | -31.56 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
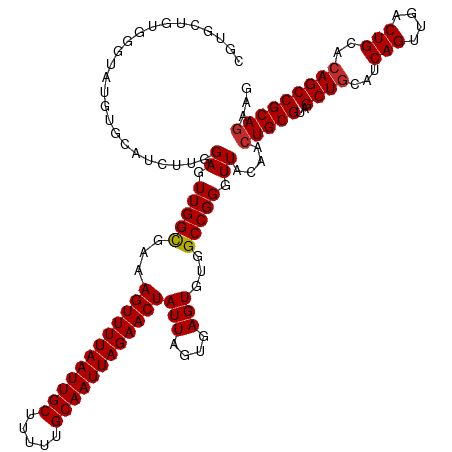
>dm3.chr3L 13721983 127 - 24543557 CGUGCUGUGGGUAUGUGCAUCUUGGAGUUGGCGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGCGGCCGGGUUACAACUGCGUAGCUGCAUCAGUUGACUGCACAGCCGCAGAAG ....((((((...((((((.......((..((...((((((((((((.....)))))))))))).....(((.((((((.(.((.......)).).)))))))))))..)))))))).))))))... ( -47.81, z-score = -2.67, R) >droEre2.scaffold_4784 13728321 127 - 25762168 CGUGCUGUGGGUAUGUGCAUCUUGGAGUUGGUGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGUGGCCGGGUUACAACUGCGUAGCUGCAUCAGUUGACUGCACAGCCGCAGAAG ....((((((...((((((.((((((.(((.(((.((((((((((((.....)))))))))))).))).))).)....)))))...((((((.((....))..))))))..)))))).))))))... ( -43.60, z-score = -1.90, R) >droYak2.chr3L 13819095 127 - 24197627 CGUGCUGUGGGUAUGUGCAUCUUGGAGUUGGUGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGUGGCCGGGUUACAACUGCGUAGCUGCAUCAGUUGACUGCACAGCCGCAGAAG ....((((((...((((((.((((((.(((.(((.((((((((((((.....)))))))))))).))).))).)....)))))...((((((.((....))..))))))..)))))).))))))... ( -43.60, z-score = -1.90, R) >droSec1.super_0 5884017 127 - 21120651 CGUUCUGUGGGUAUGUGCAUCUUGGAGUUGGCGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGUGGCCGGGUUACAACUGCGUAGCUGCAUCAGUUGACUGCACAGCCGCAGAAG ..((((((((...((((((.......((..((...((((((((((((.....)))))))))))).....(((.((..((.(.((.......)).).))..)))))))..)))))))).)))))))). ( -47.81, z-score = -3.33, R) >droSim1.chr3L 13107726 127 - 22553184 CGUGCUGUGGGUAUGUGCAUCUUGGAGUUGGCGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGUGGCCGGGUUACAACUGCGUAGCUGCAUCAGUUGACUGCACAGCCGCAGAAG ....((((((...((((((.......((..((...((((((((((((.....)))))))))))).....(((.((..((.(.((.......)).).))..)))))))..)))))))).))))))... ( -45.31, z-score = -2.17, R) >dp4.chrXR_group8 2512714 125 + 9212921 -AUUCCUUGGAUGUGUGCAUCUU-GAGUUGGUGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGUGGCCGGGUUACAACUGCGUUGCUGCAUCAGUUGACUGAACAGCCGCAGAGG -.(((...(((((....))))).-)))(((.(((.((((((((((((.....)))))))))))).))).))).((((((...)))))).(((((..((((..((((....)))).)))))))))... ( -40.20, z-score = -1.99, R) >droPer1.super_36 608422 125 - 818889 -AUUCCUUGGAUGUGUGCAUCUU-GAGUUGGUGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGUGGCCGGGUUACAACUGCGUUGCUGCAUCAGUUGACUGAACAGCCGCAGAGG -.(((...(((((....))))).-)))(((.(((.((((((((((((.....)))))))))))).))).))).((((((...)))))).(((((..((((..((((....)))).)))))))))... ( -40.20, z-score = -1.99, R) >droAna3.scaffold_13337 1594959 126 + 23293914 -UGCCCGGGUAUAUGUGCAUCUUGGAGUUGGCGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUAAGUGCGGCCGGGUUACAAAUGCGUAGCUGCAUCAGUUGACUGCAGAGCCGCAGAAG -(((....)))..((((..(((.(.(((..((...((((((((((((.....))))))))))))........(((((((.(.((.......)).).)))))))..))..))).))))..)))).... ( -41.40, z-score = -1.64, R) >droVir3.scaffold_13049 5568648 125 + 25233164 UGAGUGGUGUGUGUGUCUGUCUU-GAGUUGGCGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGC-GCCGGUUUACAACUGCGUCGCUGCCUCAGUUGACUGCACAGCCGCAGCAG ...(((((.((((((((....((-(((..(((((.((((((((((((.....))))))))))))...........(-((.(((.....)))))))))))..)))))..))).))))))))))..... ( -46.40, z-score = -2.52, R) >consensus CGUGCUGUGGGUAUGUGCAUCUUGGAGUUGGCGAAAGUUUUAAUUGCUUUUUGCAAUUAGAACUAUUAGUGAGUGUGGCCGGGUUACAACUGCGUAGCUGCAUCAGUUGACUGCACAGCCGCAGAAG ........................((.(((((...((((((((((((.....))))))))))))(((....)))...))))).))....(((((..((((...(((....)))..)))))))))... (-31.58 = -31.56 + -0.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:15 2011