| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,155,326 – 5,155,446 |
| Length | 120 |
| Max. P | 0.522452 |

| Location | 5,155,326 – 5,155,446 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.37 |
| Shannon entropy | 0.44901 |
| G+C content | 0.54583 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -21.43 |
| Energy contribution | -21.06 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.522452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5155326 120 - 23011544 AGCGGUUUCUUCUGCUGCUGCGAGGAGUCCAGUCCCUGCUUCGUCUUUUCGUGCCAGGAGCGUAUAUCCUUAAUCUUGCGUUCACAUUGGUCCCAUUCCUCGCAGGCCUCUUGCAGCAGG ...........((((((((((((((((((((((..(((...((......))...)))(((((((............)))))))..)))))....))))))))))........))))))). ( -40.60, z-score = -2.22, R) >droSim1.chr2L 4962880 120 - 22036055 AGCGGUUUCUUCUGCUGCUGCGAGGAGUCCAGUCCCUGCUUCGUCUUUUCGUGCCAGGAGCGUAUAUCCUUAAUCUUGCGUUCACAUUGGUCCCAUUCCUCGCAGGCCUCUUGCAGCAGG ...........((((((((((((((((((((((..(((...((......))...)))(((((((............)))))))..)))))....))))))))))........))))))). ( -40.60, z-score = -2.22, R) >droSec1.super_5 3241756 120 - 5866729 AGCGGUUUCUUCUGCUGCUGCGAGGAGUCCAGUCCCUGCUUCGUCUUUUCGUGCCAGGAGCGUAUAUCCUUAAUCUUGCGUUCACAUUGGUCCCAUUCCUCGCAGGCCUCUUGCAGCAGG ...........((((((((((((((((((((((..(((...((......))...)))(((((((............)))))))..)))))....))))))))))........))))))). ( -40.60, z-score = -2.22, R) >droYak2.chr2L 11955508 120 + 22324452 AGCGGUUUCUUCUGCUGCUGCGAGGAGUCCAGUCCCUGCUUCGUCUUUUCGUGCCAGGAGCGUAUAUCCUUAAUCUUGCGUUCACAUUGGUCCCAUUCCUCGCAGGCCUCUUGCAGCAGG ...........((((((((((((((((((((((..(((...((......))...)))(((((((............)))))))..)))))....))))))))))........))))))). ( -40.60, z-score = -2.22, R) >droEre2.scaffold_4929 5236468 120 - 26641161 AGCGGUUUCUUCUGCUGCUGCGAGGAGUCCAGUCCCUGCUUCGUCUUUUCGUGCCAGGAGCGUAUAUCCUUAAUCUUGCGUUCACAUUGGUCCCACUCCUCGCAGGCCUCUUGCAGCAGG ...........((((((((((((((((((((((..(((...((......))...)))(((((((............)))))))..)))))....))))))))))........))))))). ( -42.90, z-score = -2.74, R) >droAna3.scaffold_12916 6175152 120 + 16180835 AGCGGUUUCUUCUGCUGCUGCGUGGAGUCCAGGCUCUGCUUCGUCUUUUCGUGCCAGGCGCGUAUAUCCUUAAUCUUGCGUUCACAUUGAUCCCACUCUUCGCAGGCCUCUUGCAGCAGA (((((......)))))((((((..((((....))))..)..........(((((...)))))............((((((.......((....)).....))))))......)))))... ( -33.80, z-score = 0.29, R) >dp4.chr4_group3 8949426 120 + 11692001 AGCGGUUUCUUCUGCUGCUGCGACGAGUCGAGGCCCUGUUUGGUCUUCUCGUGCCAGGCGCGUAUGUCCUUGAUUUUGCGUUCGCAUUGAUCCCAUUCCUCGCAGGCCUCUUGCAGCAGA (((((......)))))(((((((....))((((((..(((((((........)))))))(((.........((((.(((....)))..))))........))).))))))..)))))... ( -44.33, z-score = -1.69, R) >droPer1.super_1 6054991 120 + 10282868 AGCGGUUUCUUCUGCUGCUGCGACGAGUCCAGGCCCUGUUUGGUCUUCUCGUGCCAGGCGCGUAUGUCCUUGAUUUUGCGUUCGCAUUGAUCCCAUUCCUCGCAGGCCUCUUGCAGCAGA (((((......))))).((((..((((...(((((..(((((((........)))))))(((.........((((.(((....)))..))))........))).)))))))))..)))). ( -40.73, z-score = -0.97, R) >droWil1.scaffold_180772 7782753 120 + 8906247 AGGGGUUUCUUAUGUUGGGGCGAAGAGUCGAGGGUUUGCUUGGUUUUCUCAUGCCAAGCGCGUAUAUCCUUAAUCUUACGUUCACAUUGGUCCCAUUCUUCGCAGGCCUCUUGCAGCAGG ..(((...((.((((..(((((((((...((((((.((((((((........)))))))).....))))))..)))).))))))))).)).))).......(((((...)))))...... ( -38.10, z-score = -1.52, R) >droVir3.scaffold_12963 13764970 120 + 20206255 AGCGGCUUCUUUUGCUGCGGCGAGGAGUCCAAUGUUUGUUUGGUUUUCUCAUGCCAGGCGCGUAUGUCCUUGAUCUUGCGCUCGCAUUGAUCCCACUCCUCGCAGGCCUCCUGCAGCAGU .(((((.......))))).(((((((((.((((((..(((((((........)))))))(((((.(((...)))..)))))..)))))).....)))))))(((((...))))).))... ( -48.20, z-score = -2.77, R) >droMoj3.scaffold_6500 6765603 120 + 32352404 AGCGGCUUCUUUUGCUGUGGCGAGGAGUCUAAGGUCUGUUUGGUCUUCUCGUGCCAGGCACGAAUAUCCUUGAUCUUGCGCUCACAUUGAUCCCACUCUUCACAGGCCUCCUGCAGCAGC ...........((((((..(.(((((((.(((((((.....((.....((((((...))))))....))..))))))).))))....(((.........))).....))))..)))))). ( -36.50, z-score = 0.16, R) >droGri2.scaffold_15252 7522528 120 + 17193109 AGCGGCUUCUUUAGCUGCGGCGAUGAGUCCAAUGUUUGUUUGGUCUUUUCAUGCCAGGCGCGUAUGUCCUUCACCUUGCGCUCGCAUUGAUCCCACUCCUCGCAGGCCUCCUGCAGCAGC .((((((.....)))))).((((.((((.((((((..(((((((........)))))))(((((.((.....))..)))))..)))))).....)))).))(((((...))))).))... ( -41.40, z-score = -1.39, R) >anoGam1.chr3R 16620711 120 + 53272125 AACGGUUUGCUCUUCAGCUCGCCAGAAUUCAAGGCCAGAUAGGCGUCCUUUAUCCACUCCCGUACGCUCUCGAUCUUCCGCUCGUACUGCUGCCACUCUUCGGUCGUUUCGUGCAGGAAC ...(((..(((....)))..)))..........(((.....))).............(((.(((((....(((((........(((....)))........)))))...))))).))).. ( -25.49, z-score = 1.37, R) >triCas2.chrUn_44 212439 120 - 525920 AGUGGCUUCUUCUUGUUCUGUUGAGAGUCCAGAGUGGUUGUGGUUUUUUCGAUCCAACCGCGGACGUCCUUCAUCUUCUUCUCGCACUGUUCCCACUCUUCGGACGUUUCUUGGAGCAAA ............((((((((..(((((((((((((((..((((((..........))))))((....)).......................)))))))..)))).)))).)))))))). ( -33.30, z-score = -0.65, R) >consensus AGCGGUUUCUUCUGCUGCUGCGAGGAGUCCAGUCCCUGCUUGGUCUUUUCGUGCCAGGCGCGUAUAUCCUUAAUCUUGCGUUCACAUUGAUCCCACUCCUCGCAGGCCUCUUGCAGCAGG ...........(((((((.(((((((((....(((..((.............))..)))(((((............))))).............)))))))))(((...)))))))))). (-21.43 = -21.06 + -0.38)


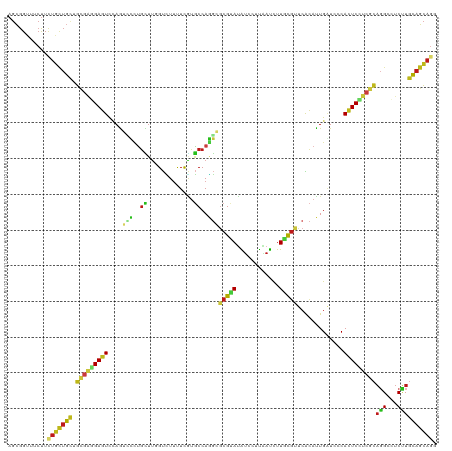
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:14 2011