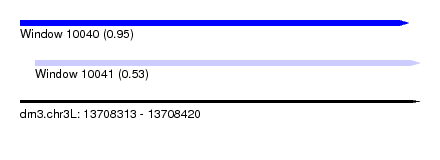
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,708,313 – 13,708,420 |
| Length | 107 |
| Max. P | 0.953467 |

| Location | 13,708,313 – 13,708,417 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.24 |
| Shannon entropy | 0.49033 |
| G+C content | 0.38802 |
| Mean single sequence MFE | -18.16 |
| Consensus MFE | -11.62 |
| Energy contribution | -11.07 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13708313 104 + 24543557 UUGGGUUGACACCUUGAGUGUCAUUAAUGCGUGUGACAUUUUCAUUAUAACUUUUUAUUGUUCUCC--UUUCUCGCUUUCCUUUCCUUCGCCC---AAAUCCCCUAAGU ((((((((((((.....)))))).....(((.(.........((.((........)).))......--...).))).............))))---))........... ( -17.67, z-score = -2.05, R) >droEre2.scaffold_4784 13714188 104 + 25762168 UUGGGUUGACACCUUGAGUGUCAUUAAUGCGUGUGACAUUUUCAUUAUAACUUUUUAUUGUUCUCU--UUUCUCGCCUUCCUUUCCGUCGCCC---AAAUCCCCAAAGU ((((((.(((.....(((((((((........))))))))).......(((........)))....--..................)))))))---))........... ( -19.80, z-score = -2.31, R) >droYak2.chr3L 13804888 104 + 24197627 UUGGGUUGACACCUUGAGUGUCAUUAAUGCGUGUGACAUUUUCAUUAUAACUUUUUAUUGUUCUCU--UUUCUCGCUUUCCUUUCCUUCGCCC---AAAUCCCACAAGU ((((((((((((.....)))))).....(((.(.........((.((........)).))......--...).))).............))))---))........... ( -17.67, z-score = -1.68, R) >droSec1.super_0 5870481 104 + 21120651 UUGGGUUGACACCUUGAGUGUCAUUAAUGCGUGUGACAUUUUCAUUAUAACUUUUUAUUGUUCUCU--UUUCUCGCUUUCCUUUCCUUCGCCC---AAAUCCCCUAAGU ((((((((((((.....)))))).....(((.(.........((.((........)).))......--...).))).............))))---))........... ( -17.67, z-score = -2.07, R) >droSim1.chr3L 13094133 104 + 22553184 UUGGGUUGACACCUUGAGUGUCAUUAAUGCGUGUGACAUUUUCAUUAUAACUUUUUAUUGUUCUCU--UUUCUCGCUUUCCUUUCCUUCGCCC---AAAUCCCCUAAGU ((((((((((((.....)))))).....(((.(.........((.((........)).))......--...).))).............))))---))........... ( -17.67, z-score = -2.07, R) >droWil1.scaffold_180698 10958049 108 - 11422946 -CGAGUUGACACCUUGAGUGUCAUUAAUGCGUAUGACAUUUUCAUUAUAAGUUUUUAUUGUUCUCUACCUUGUACAUUUCCUUUUCAAUACCCUGUAAUCCAUUCAAGU -.((((.((......(((((((((........)))))))))....((((((.................))))))........................)).)))).... ( -12.43, z-score = 0.14, R) >droMoj3.scaffold_6680 17380556 99 - 24764193 --GAGCUGUCACCUUGAGUGUCAUUAAUGCAUGUGACAUUUUCAUUAUAGCUACUC--U-UUCUCUCUGGGAUUUCUCGCCUUUUUGGGCUGUCCAAAUUUAAC----- --.((((((......(((((((((........))))))))).....))))))....--.-.........((((.....((((....)))).)))).........----- ( -24.30, z-score = -2.36, R) >droVir3.scaffold_13049 5555967 94 - 25233164 --GGGCUGUCACCUUGAGUGUCAUUAAUGCAAGUGACAUUUUCAUUAUAGCUACUCGCU-UUCUCUUACGGAUUUCUCGCAUUUUCAG-CUGCCAAAA----------- --(((((((......((((((((((......)))))))))).....))))).....(((-........(((.....))).......))-)..))....----------- ( -18.06, z-score = -0.40, R) >consensus UUGGGUUGACACCUUGAGUGUCAUUAAUGCGUGUGACAUUUUCAUUAUAACUUUUUAUUGUUCUCU__UUUCUCGCUUUCCUUUCCUUCGCCC___AAAUCCCCUAAGU ...(((((.......(((((((((........)))))))))......)))))......................................................... (-11.62 = -11.07 + -0.55)

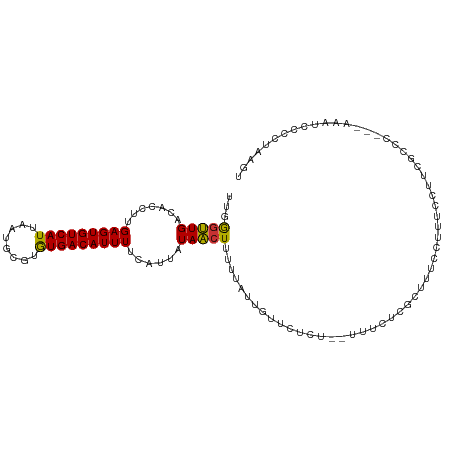
| Location | 13,708,317 – 13,708,420 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.99 |
| Shannon entropy | 0.50207 |
| G+C content | 0.39144 |
| Mean single sequence MFE | -14.88 |
| Consensus MFE | -10.43 |
| Energy contribution | -10.07 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.56 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13708317 103 + 24543557 GUUGACACCUUGAGUGUCAUUAAUGCGUGUGACAUUUUCAUUAUAACUUUUUAUUGUUCUCC--UUUCUCGCUUUCCUUUCCUUCGCCC---AAAUCCCCUAAGUGGG ((((.......(((((((((........)))))))))......))))...............--...(((((((...............---.........))))))) ( -13.98, z-score = -0.76, R) >droEre2.scaffold_4784 13714192 103 + 25762168 GUUGACACCUUGAGUGUCAUUAAUGCGUGUGACAUUUUCAUUAUAACUUUUUAUUGUUCUCU--UUUCUCGCCUUCCUUUCCGUCGCCC---AAAUCCCCAAAGUGGG ...(((.....(((((((((........))))))))).......(((........)))....--..................)))....---.....(((.....))) ( -14.60, z-score = -0.48, R) >droYak2.chr3L 13804892 103 + 24197627 GUUGACACCUUGAGUGUCAUUAAUGCGUGUGACAUUUUCAUUAUAACUUUUUAUUGUUCUCU--UUUCUCGCUUUCCUUUCCUUCGCCC---AAAUCCCACAAGUGGG ((((.......(((((((((........)))))))))......))))...............--.........................---....((((....)))) ( -14.72, z-score = -0.62, R) >droSec1.super_0 5870485 103 + 21120651 GUUGACACCUUGAGUGUCAUUAAUGCGUGUGACAUUUUCAUUAUAACUUUUUAUUGUUCUCU--UUUCUCGCUUUCCUUUCCUUCGCCC---AAAUCCCCUAAGUGGU ((((.......(((((((((........)))))))))......))))...............--....((((((...............---.........)))))). ( -11.98, z-score = -0.38, R) >droSim1.chr3L 13094137 103 + 22553184 GUUGACACCUUGAGUGUCAUUAAUGCGUGUGACAUUUUCAUUAUAACUUUUUAUUGUUCUCU--UUUCUCGCUUUCCUUUCCUUCGCCC---AAAUCCCCUAAGUGGU ((((.......(((((((((........)))))))))......))))...............--....((((((...............---.........)))))). ( -11.98, z-score = -0.38, R) >droWil1.scaffold_180698 10958052 108 - 11422946 GUUGACACCUUGAGUGUCAUUAAUGCGUAUGACAUUUUCAUUAUAAGUUUUUAUUGUUCUCUACCUUGUACAUUUCCUUUUCAAUACCCUGUAAUCCAUUCAAGUAGG .....(..((((((((..((((....((((((.........((((((.................))))))..........)).))))....)))).))))))))..). ( -12.24, z-score = 0.36, R) >droMoj3.scaffold_6680 17380558 101 - 24764193 GCUGUCACCUUGAGUGUCAUUAAUGCAUGUGACAUUUUCAUUAUAGCUACUC--U-UUCUCUCUGGGAUUUCUCGCCUUUUUGGGCUGUCCAAAUUUAACAGAA---- (((((......(((((((((........))))))))).....))))).....--.-.....((((((((.....((((....)))).))))........)))).---- ( -23.40, z-score = -1.83, R) >droVir3.scaffold_13049 5555969 92 - 25233164 GCUGUCACCUUGAGUGUCAUUAAUGCAAGUGACAUUUUCAUUAUAGCUACUCGCU-UUCUCUUACGGAUUUCUCGCAUUUUCAG-CUGCCAAAA-------------- (((((......((((((((((......)))))))))).....))))).....(((-........(((.....))).......))-)........-------------- ( -16.16, z-score = -0.42, R) >consensus GUUGACACCUUGAGUGUCAUUAAUGCGUGUGACAUUUUCAUUAUAACUUUUUAUUGUUCUCU__UUUCUCGCUUUCCUUUCCUUCGCCC___AAAUCCCCUAAGUGGG ((((.......(((((((((........)))))))))......))))............................................................. (-10.43 = -10.07 + -0.36)
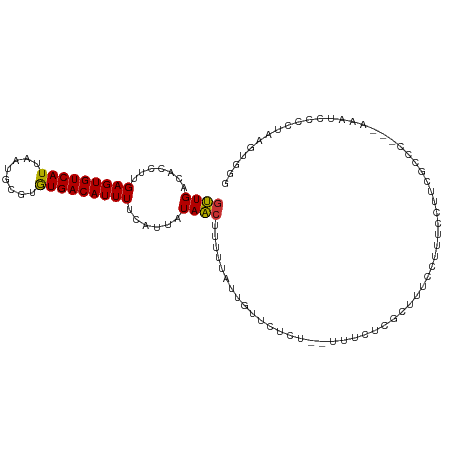
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:09 2011