| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,696,888 – 13,696,979 |
| Length | 91 |
| Max. P | 0.936161 |

| Location | 13,696,888 – 13,696,979 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 80.78 |
| Shannon entropy | 0.38345 |
| G+C content | 0.31093 |
| Mean single sequence MFE | -11.19 |
| Consensus MFE | -8.95 |
| Energy contribution | -9.08 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13696888 91 + 24543557 UAUAAAUAAGUAAUAAAUACGCUUAGUUGCAUCAAAAAGUGCCACUAACCGAAAAAUAGUUGGCCAACCAAAAAACAAAAAAAAAGAGAGA ...................((.(((((.((((......)))).))))).))........((((....)))).................... ( -12.30, z-score = -2.37, R) >droSim1.chr3L 13082509 82 + 22553184 UAUAAAUAAGUAAUAAAUACGCUUAGUUGCAUCAAAAAGUGCCACUAACCGAAAAAUAGUUGGCCAACCCGAAAAAAAGGGA--------- ...................((.(((((.((((......)))).))))).))................(((........))).--------- ( -14.80, z-score = -1.89, R) >droSec1.super_0 5859011 82 + 21120651 UAUAAAUAAGUAAUAAAUACGCUUAGUUGCAUCAAAAAGUGCCACUAACCGAAAAAUAGUUGGCCAACCCGAAAAAAAGAGA--------- ...................((.(((((.((((......)))).))))).))........((((.....))))..........--------- ( -11.60, z-score = -2.00, R) >droYak2.chr3L 13793147 82 + 24197627 UAUAAAUAAGUAAUAAAUACGCUUAGUUGCAUCAAAAAGUGCCACUAACCGAAAAAUAGUUGGCCAACCCAAAAAAAACAAA--------- ...................((.(((((.((((......)))).))))).))........((((.....))))..........--------- ( -11.90, z-score = -2.32, R) >dp4.chrXR_group8 2485471 69 - 9212921 UAUAAAUAAGUAAUAAAUACGCUUAGUUGCAUCAAAAAGUGCCACUAACCGAAAAAUAGUUGCCCAACA---------------------- .........(((((.....((.(((((.((((......)))).))))).)).......)))))......---------------------- ( -10.80, z-score = -2.40, R) >droPer1.super_36 581054 69 + 818889 UAUAAAUAAGUAAUAAAUACGCUUAGUUGCAUCAAAAAGUGCCACUAACCGAAAAAUAGUUGCCCAACA---------------------- .........(((((.....((.(((((.((((......)))).))))).)).......)))))......---------------------- ( -10.80, z-score = -2.40, R) >droMoj3.scaffold_6680 17362890 88 - 24764193 UAUAAAUAAGUAAUAAAUACGCUUAGUUGCAUCGAGAAGUGCCACUAGCCGAAAAAUAGUUGCCAGACAAAUAAAU---ACAAGAGAUAAA .........(((.......((.(((((.((((......)))).))))).)).......(((....))).......)---)).......... ( -11.70, z-score = -1.43, R) >droGri2.scaffold_15110 17660798 91 + 24565398 UAUAAAUAAGUAAUAAAUACGCUUAGUUGCAUCAAAAAGUGGCACUAACCGAAAAAUAGUUGGCAGAAAAAUAUAUACUACAACAGAUAAA ...................((.(((((.((..(.....)..))))))).)).......((((..................))))....... ( -8.47, z-score = 0.39, R) >droVir3.scaffold_13049 5539343 88 - 25233164 UAUAAAUAAGUAAUAAAUACGCUUAGUUGCAUCAAAAAGUGGCACUAACCGAAAAAUAGUUGGCAGACAAACAUAU---ACAAGAGAUAAA .........(((.......((.(((((.((..(.....)..))))))).)).......(((........)))...)---)).......... ( -8.30, z-score = 0.37, R) >consensus UAUAAAUAAGUAAUAAAUACGCUUAGUUGCAUCAAAAAGUGCCACUAACCGAAAAAUAGUUGGCCAACAAAAAAAAA__ACA_________ ...................((.(((((.((((......)))).))))).))........................................ ( -8.95 = -9.08 + 0.12)

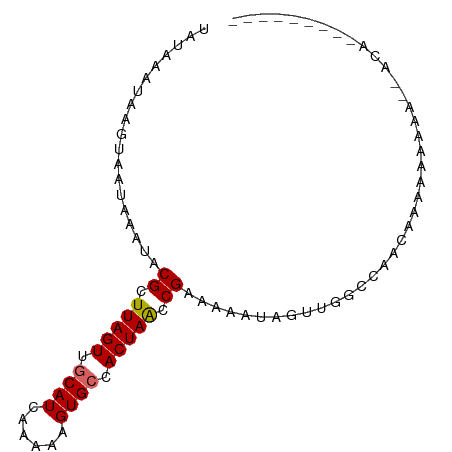
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:06 2011