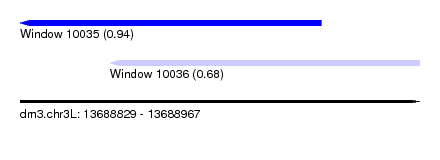
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,688,829 – 13,688,967 |
| Length | 138 |
| Max. P | 0.939022 |

| Location | 13,688,829 – 13,688,933 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.86 |
| Shannon entropy | 0.57073 |
| G+C content | 0.47632 |
| Mean single sequence MFE | -27.21 |
| Consensus MFE | -13.16 |
| Energy contribution | -13.25 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
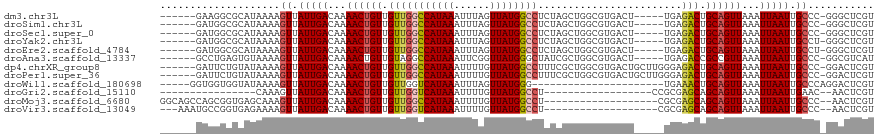
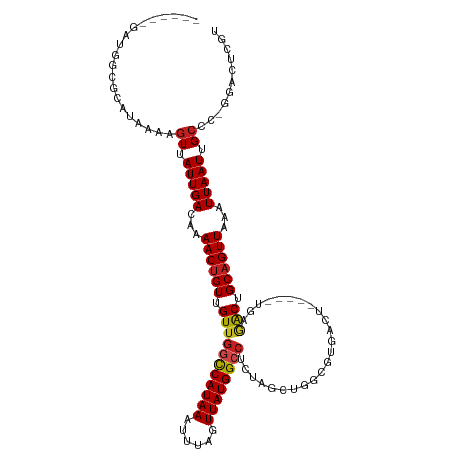
>dm3.chr3L 13688829 104 - 24543557 UGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACU-----UGAGACUGCAGUUAAAUUAAUUGCCCG-GGCUCGUAAACACCU---CCGAAAGUCCUCAACCGCCACUC--- .((((((((......))))))))...((.(((((.(..(-----((((((((((((((...))))))).((-((...(....)...)---)))..))).))))))))))))).--- ( -32.00, z-score = -2.63, R) >droSim1.chr3L 13074083 104 - 22553184 UGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACU-----UGAGACUGCAGUUAAAUUAAUUGCCCG-GGCUCGUAAACACCU---CCGAAAGUCCUCAACCACAACUC--- .((((((((......))))))))..........(((..(-----((((((((((((((...))))))).((-((...(....)...)---)))..))).))))).))).....--- ( -27.60, z-score = -1.64, R) >droSec1.super_0 5851037 104 - 21120651 UGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACU-----UGAGACUGCAGUUAAAUUAAUUGCCCG-GGCUCGUAAACACCU---CCGAAAGUCCUCAACCACCACUC--- .((((((((......))))))))........(.(((..(-----((((((((((((((...))))))).((-((...(....)...)---)))..))).))))).))))....--- ( -26.80, z-score = -1.29, R) >droYak2.chr3L 13784921 104 - 24197627 UGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACU-----UGAGACUGCAGUUAAAUUAAUUGCCUG-GGCUCGUAAACACCU---CCGAAAGUCCUCGACCACCACUC--- .((((((((......))))))))....(.(((((.((((-----((((.(((((((((...)))))))..)-).)))(....)....---....)))))..)).))).)....--- ( -28.40, z-score = -1.61, R) >droEre2.scaffold_4784 13694318 104 - 25762168 UGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACU-----UGAGACUGCAGUUAAAUUAAUUGCCUG-GGCUCGUAAACACCU---CCGAAAGUCCUCAACCACCGCUC--- .((((((((......)))))))).......((((((..(-----((((((((((((((...)))))))..(-(...........)).---.....))).))))).)).)))).--- ( -27.60, z-score = -1.23, R) >droAna3.scaffold_13337 21745959 101 - 23293914 AGGCCAUAAAUUCGGUUAUGGGCUAUCGCUGGCGUGACU-----UGAGACCGCCGUUAAAUUAAUUGCCCG-GCGUCAUAAACACCU---CCGCAAGUUUUGUAUCCUUC------ ((.((((((......)))))).))...((.((.(((..(-----((.(((.((((..............))-))))).))).))).)---).))................------ ( -21.94, z-score = 0.95, R) >dp4.chrXR_group8 6738048 102 - 9212921 UGGCCAUAAAUUUUGUUAUGGCCUUUCGCUGGCGUGACUGCUUGGGAGACUGCAGUUAAAUUAAUUGCCCG-GACUCGUAAACACCU---CC-CAAGCACCCCAUCC--------- .((((((((......))))))))..((((....)))).((((((((((...(((((((...))))))).((-....)).......))---))-))))))........--------- ( -35.60, z-score = -3.59, R) >droPer1.super_36 248442 102 + 818889 UGGCCAUAAAUUUUGUUAUGGCCUUUCGCUGGCGUGACUGCUUGGGAGACUGCAGUUAAAUUAAUUGCCCG-GACUCGUAAACACCU---CC-CAAACACCCCAUCC--------- .((((((((......))))))))......(((.(((.....(((((((...(((((((...))))))).((-....)).......))---))-))).))).)))...--------- ( -30.30, z-score = -2.38, R) >droWil1.scaffold_180698 502704 91 - 11422946 UGGUCAUAAAUUUAGUUAUGGGUG----------------------AAACUGCAGUUAAAUUAAUUGCCCAGGACUCGUAAACACCUAUAGCAUUAGG-CCGCAACACCCCCUC-- .((((.(((.....((((((((((----------------------.....(((((((...)))))))...(....).....)))))))))).)))))-)).............-- ( -19.80, z-score = -0.46, R) >droGri2.scaffold_15110 6915942 96 + 24565398 UGGUCAUAAAUUUUGUUAUGGCCUCC------------------GCGAGCAGCAGUUAAAUUAAUUGAACA--ACUCGUAAACACCUUCUCCCCAAGGAGCGCGCAACACCCACGC .((((((((......))))))))...------------------(((.((..((((((...))))))....--...........((((......)))).)).)))........... ( -21.40, z-score = -1.12, R) >droVir3.scaffold_13049 19704856 95 - 25233164 UGGUCAUAAAUUUUGUUAUGGCCU-C------------------GCGAGCAGCAGUUAAAUUAAUUGCCCA--ACUCGUAAACACCUUCUCUCCAAGGAGAGCAGAACCCACACAC .((((((((......)))))))).-.------------------(((((..(((((((...)))))))...--.)))))......((.(((((....))))).))........... ( -27.90, z-score = -3.84, R) >consensus UGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACU_____UGAGACUGCAGUUAAAUUAAUUGCCCG_GACUCGUAAACACCU___CCGAAAGUCCUCCACCACCACUC___ .((((((((......))))))))............................(((((((...)))))))................................................ (-13.16 = -13.25 + 0.08)
| Location | 13,688,860 – 13,688,967 |
|---|---|
| Length | 107 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Shannon entropy | 0.46132 |
| G+C content | 0.41407 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -16.59 |
| Energy contribution | -16.60 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.683798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
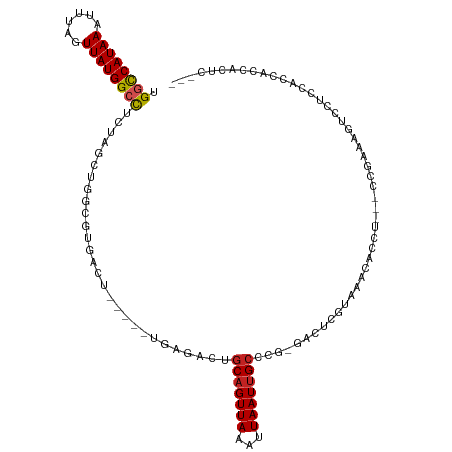
>dm3.chr3L 13688860 107 - 24543557 ------GAAGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACU-----UGAGACUGCAGUUAAAUUAAUUGCCC-GGGCUCGU ------....((((.....(((((..(((((.....)))))((((((((......))))))))..))))).))))....-----.(((.(((((((((...)))))))..-)).))).. ( -29.70, z-score = -1.03, R) >droSim1.chr3L 13074114 107 - 22553184 ------GAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACU-----UGAGACUGCAGUUAAAUUAAUUGCCC-GGGCUCGU ------..(.((((.....(((((..(((((.....)))))((((((((......))))))))..))))).)))).)..-----.(((.(((((((((...)))))))..-)).))).. ( -29.80, z-score = -1.08, R) >droSec1.super_0 5851068 107 - 21120651 ------GAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACU-----UGAGACUGCAGUUAAAUUAAUUGCCC-GGGCUCGU ------..(.((((.....(((((..(((((.....)))))((((((((......))))))))..))))).)))).)..-----.(((.(((((((((...)))))))..-)).))).. ( -29.80, z-score = -1.08, R) >droYak2.chr3L 13784952 107 - 24197627 ------GAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACU-----UGAGACUGCAGUUAAAUUAAUUGCCU-GGGCUCGU ------..(.((((.....(((((..(((((.....)))))((((((((......))))))))..))))).)))).)..-----.(((.(((((((((...)))))))..-)).))).. ( -29.80, z-score = -1.22, R) >droEre2.scaffold_4784 13694349 107 - 25762168 ------GAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACU-----UGAGACUGCAGUUAAAUUAAUUGCCU-GGGCUCGU ------..(.((((.....(((((..(((((.....)))))((((((((......))))))))..))))).)))).)..-----.(((.(((((((((...)))))))..-)).))).. ( -29.80, z-score = -1.22, R) >droAna3.scaffold_13337 21745987 107 - 23293914 ------GCCUGAGUGUAAAAGUUAUUGACAAAACUGUUGUAGGCCAUAAAUUCGGUUAUGGGCUAUCGCUGGCGUGACU-----UGAGACCGCCGUUAAAUUAAUUGCCC-GGCGUCAU ------(((.(.(.((((......(((((......((.((((.((((((......)))))).)))).)).((((.(...-----.....)))))))))).....))))))-)))..... ( -27.00, z-score = 0.03, R) >dp4.chrXR_group8 6738072 112 - 9212921 ------GAUUCUGUAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUUGUUAUGGCCUUUCGCUGGCGUGACUGCUUGGGAGACUGCAGUUAAAUUAAUUGCCC-GGACUCGU ------((.((((......(((....(((((.....)))))((((((((......))))))))....)))(((((((((((..((....))))))))).......)))))-))).)).. ( -32.11, z-score = -2.38, R) >droPer1.super_36 248466 112 + 818889 ------GAUUCUGUAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUUGUUAUGGCCUUUCGCUGGCGUGACUGCUUGGGAGACUGCAGUUAAAUUAAUUGCCC-GGACUCGU ------((.((((......(((....(((((.....)))))((((((((......))))))))....)))(((((((((((..((....))))))))).......)))))-))).)).. ( -32.11, z-score = -2.38, R) >droWil1.scaffold_180698 502738 92 - 11422946 -----GGUGGUGGUAUAAAAGUUAUUGACAAAACUGUUGUUGGUCAUAAAUUUAGUUAUGGG----------------------UGAAACUGCAGUUAAAUUAAUUGCCCAGGACUCGU -----.(.((..((......(((...)))..((((((..(((.((((((......)))))).----------------------)))....))))))......))..)))......... ( -15.80, z-score = 0.56, R) >droGri2.scaffold_15110 6915979 83 + 24565398 ----------------CAAAGUUAUUGACAAAACUGUUGUUGGUCAUAAAUUUUGUUAUGGCCU------------------CCGCGAGCAGCAGUUAAAUUAAUUGAAC--AACUCGU ----------------....((((((((...((((((((((((((((((......)))))))).------------------.....))))))))))...)))))..)))--....... ( -20.40, z-score = -2.10, R) >droMoj3.scaffold_6680 7411132 98 - 24764193 GGCAGCCAGCGGUGAGCAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUUGUUAUGGCCU-------------------CGCGAGCAGCAGUUAAAUUAAUUGCCC--AACUCGU (((((...((.....))..............((((((((((((((((((......)))))))).-------------------....)))))))))).......))))).--....... ( -29.70, z-score = -1.55, R) >droVir3.scaffold_13049 19704893 95 - 25233164 ---AAAUGCCGGUGAGAAAAGUUAUUGACAAAACUGUUGUUGGUCAUAAAUUUUGUUAUGGCCU-------------------CGCGAGCAGCAGUUAAAUUAAUUGCCC--AACUCGU ---..........(((....((.(((((...((((((((((((((((((......)))))))).-------------------....))))))))))...))))).))..--..))).. ( -24.30, z-score = -1.70, R) >consensus ______GAUGGCGCAUAAAAGUUAUUGACAAAACUGUUGUUGGCCAUAAAUUUAGUUAUGGCCUCUAGCUGGCGUGACU_____UGAGACUGCAGUUAAAUUAAUUGCCC_GGACUCGU ....................((.(((((...((((((.(((((((((((......))))))))....(....)..............))).))))))...))))).))........... (-16.59 = -16.60 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:05 2011