
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,688,608 – 13,688,689 |
| Length | 81 |
| Max. P | 0.808455 |

| Location | 13,688,608 – 13,688,689 |
|---|---|
| Length | 81 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 56.64 |
| Shannon entropy | 0.78304 |
| G+C content | 0.38371 |
| Mean single sequence MFE | -18.42 |
| Consensus MFE | -4.85 |
| Energy contribution | -4.72 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.808455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
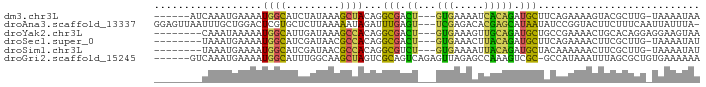
>dm3.chr3L 13688608 81 - 24543557 ------AUCAAAUGAAAAUGGCAUCUAUAAAGCUACAGGCGACU---GUGAAAAUCACAGAUGCUUCAGAAAAGUACGCUUG-UAAAAUAA ------...........................((((((((.((---(((.....))))).(((((.....)))))))))))-))...... ( -19.50, z-score = -2.92, R) >droAna3.scaffold_13337 21745715 87 - 23293914 GGAGUUAAUUUGCUGGACUCGUGCUCUUAAAAAUAGAUUUGAGU---UCGAGACACGAGCAUAAUAUCCGGUACUUCUUUCAAUUAUUUA- (((((......((((((((((((((((((((......)))))..---..))).)))))).......))))))))))).............- ( -19.71, z-score = -1.59, R) >droYak2.chr3L 13784700 80 - 24197627 --------CAAAUAAAAAUGGCAUUGAUAAAGCCACAGGCGACU---GUGAAAGUUGCAGAUGCUGCCGAAAACUGCACAGGAGGAAGUAA --------..........((((.........))))...((((((---.....))))))...((((.((.....((....))..)).)))). ( -16.70, z-score = -0.59, R) >droSec1.super_0 5850819 79 - 21120651 --------UAAAUGAAAAUGGCAUCGAUAACGCCACAGGCGACU---GUGAAACUUACAGAUGCUUCAGAAAACUUCGCUUG-UAAAAUAU --------....(((....((((((..(((...(((((....))---)))....)))..)))))))))..............-........ ( -16.40, z-score = -1.97, R) >droSim1.chr3L 13073865 79 - 22553184 --------UAAAUGAAAAUGGCAUCGAUAACGCCACAGGCGUCU---GUGAAAAUUACAGAUGCUACAAAAAACUUCGCUUG-UAAAAUAU --------.....(((..((((.........))))..(((((((---((((...))))))))))).........))).....-........ ( -18.70, z-score = -3.18, R) >droGri2.scaffold_15245 17879589 84 - 18325388 ------GUCAAAUGAAAAUGGCAUUUGGCAAGCUAGUCGCAGUCAGAGUUAGAGCCAAAGUCGC-GCCAUAAAUUUAGCGCUGUGAAAAAA ------.(((.........(((.((((((...((((((.......)).)))).)))))))))((-((.(......).))))..)))..... ( -19.50, z-score = -0.63, R) >consensus ________CAAAUGAAAAUGGCAUCGAUAAAGCCACAGGCGACU___GUGAAAAUCACAGAUGCUACAGAAAACUUCGCUUG_UAAAAUAA ..................(((((((......((.....)).......(((.....))).)))))))......................... ( -4.85 = -4.72 + -0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:04 2011