
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,154,775 – 5,154,874 |
| Length | 99 |
| Max. P | 0.992442 |

| Location | 5,154,775 – 5,154,874 |
|---|---|
| Length | 99 |
| Sequences | 15 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.63 |
| Shannon entropy | 0.57206 |
| G+C content | 0.55074 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -15.01 |
| Energy contribution | -14.24 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 2.05 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.992442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5154775 99 + 23011544 ---------GGUCAAGACCAACGGUCAGCUGAUCAUCGAGAACUGUCGCAACAGCGAGGAGAAGACGCUGGUGCAGACGACCAUCGAUCAGUUGGCCGCAUCGCUGGC------ ---------(((....)))..(((((((((((...((((....(((((((.(((((.........))))).))).))))....)))))))))))))))..........------ ( -39.30, z-score = -2.27, R) >droSim1.chr2L 4962333 99 + 22036055 ---------GGUCAAGACCAACGGCCAGCUGAUCAUCGAGAACUGUCGCAACAGCGAGGAGAAGACGCUGGUGCAGACGACCAUCGAUCAGUUGGCCGCAUCGCUGGC------ ---------(((....)))..(((((((((((...((((....(((((((.(((((.........))))).))).))))....)))))))))))))))..........------ ( -42.00, z-score = -3.09, R) >droSec1.super_5 3241209 99 + 5866729 ---------GGUCAAGACCAACGGCCAGCUGAUCAUCGAGAACUGUCGCAACAGCGAGGAGAAGACGCUGGUGCAGACGACCAUCGAUCAGUUGGCCGCAUCGCUGGC------ ---------(((....)))..(((((((((((...((((....(((((((.(((((.........))))).))).))))....)))))))))))))))..........------ ( -42.00, z-score = -3.09, R) >droYak2.chr2L 11954959 99 - 22324452 ---------GGUCAAGACCAACGGCCAGCUGAUCAUCGAGAACUGUCGCAACAGCGAGGAGAAGACGCUGGUGCAGACGACCAUCGAUCAGUUGGCCGCAUCGCUGGC------ ---------(((....)))..(((((((((((...((((....(((((((.(((((.........))))).))).))))....)))))))))))))))..........------ ( -42.00, z-score = -3.09, R) >droEre2.scaffold_4929 5235922 99 + 26641161 ---------GGUCAAGACCAAUGGCCAACUGAUCAUCGAGAACUGUCGCAACAGCGAGGAGAAGACGCUGGUGCAGACGACCAUCGACCAGUUGGCCGCAUCGCUGGC------ ---------(((....)))...(((((((((....((((....(((((((.(((((.........))))).))).))))....)))).)))))))))...........------ ( -38.20, z-score = -2.44, R) >droAna3.scaffold_12916 6174614 99 - 16180835 ---------GGUUAAGACCAACGGCCAGCUGAUCAUCGAGAACUGUCGCAACAGCGAGGAGAAGACGCUGGUGCAGACAACCAUCGAUCAGUUGGCCGCCUCUCUGGC------ ---------(((....)))...((((((((((...((((....(((((((.(((((.........))))).))).))))....))))))))))))))(((.....)))------ ( -44.10, z-score = -4.15, R) >dp4.chr4_group3 8948911 99 - 11692001 ---------GGUCAAGACCAACGGCCAGCUGAUUAUCGAGAAUUGCCGCAACAGUGAGGAGAAGACACUGGUGCAGACGACCAUCGAUCAGUUGGCCGCUUCCCUGGC------ ---------(((....)))..(((((((((((...((((...((((.(((.(((((.........))))).))).).)))...)))))))))))))))..........------ ( -38.10, z-score = -2.96, R) >droPer1.super_1 6054476 99 - 10282868 ---------GGUCAAGACCAACGGCCAGCUGAUUAUCGAGAAUUGCCGCAACAGUGAGGAGAAGACACUGGUGCAGACGACCAUCGAUCAGUUGGCCGCUUCCCUGGC------ ---------(((....)))..(((((((((((...((((...((((.(((.(((((.........))))).))).).)))...)))))))))))))))..........------ ( -38.10, z-score = -2.96, R) >droWil1.scaffold_180772 7782175 99 - 8906247 ---------GGUCAAAACAAAUGGACAGCUAAUUAUUGAGAAUUGUCGCAACAGUGAGGAGAAGACACUGGUGCAGACGACAAUCGAUCAAUUGGCUGCCUCAUUGGC------ ---------........(((..((.((((((((((((((...((((((((.(((((.........))))).))).)))))...))))).)))))))))))...)))..------ ( -33.90, z-score = -3.89, R) >droVir3.scaffold_12963 13764433 99 - 20206255 ---------GGUCAAGACCAACGGUCAGCUGAUCAUUGAGAACUGUCGCAACAGCGAGGAGAAGACGCUGGUGCAAUCGACCAUUGAUCAGCUGGCCGCGCAGCUAGC------ ---------(((....)))..((((((((((((((.((.((....))(((.(((((.........))))).))).......)).))))))))))))))..........------ ( -41.00, z-score = -3.21, R) >droMoj3.scaffold_6500 6765078 99 - 32352404 ---------GGUCAAGACCAACGGCCAGCUGAUCAUUGAGAAUUGUCGCAACAGCGAGGAGAAGACGCUGGUGCAGUCGACCAUUGAUCAGUUGGCCGCUCAGCUAGC------ ---------(((....)))..((((((((((((((.((....((.((((....)))).))...(((((....)).)))...)).))))))))))))))..........------ ( -42.50, z-score = -3.86, R) >droGri2.scaffold_15252 7522010 99 - 17193109 ---------GGUCAAGACCAACGGCCAGCUGAUCAUUGAGAACUGUCGCAACAGCGAGGAGAAGACGCUGGUGCAAUCGACCAUUGAUCAGUUGGCCGCCCAGCUGGC------ ---------(((....)))...(((((((((((((.((.((....))(((.(((((.........))))).))).......)).)))))))))))))(((.....)))------ ( -42.20, z-score = -3.55, R) >anoGam1.chr3R 16620053 114 - 53272125 GGCCAACAUGGAUCAGCUUACGCACGAGCUGGACAAAACGAUCGCGCUGAUCGAGCAGAAGAACCAGACGGUCGAUCAGAUCCUCGGCAAUUGGGCGAACUUUAUGCGGCUGUA ((((..(((((((((((((......))))))).........((((.((((((((.(.............).))))))))..((.........))))))..)))))).))))... ( -34.42, z-score = -0.49, R) >apiMel3.Group1 9427581 99 - 25854376 ---------AUUAACGAGAAAUGGCCGUACGAUUGCGGAGAACACGAGGGACGACACGGAGAAACAAUUGAUCGACAGCACCGUUCACAACGUGACCGAACAGUUGAA------ ---------.(((((......((((((((....)))))....((((..(((((....(..((.........))..).....)))))....)))).)))....))))).------ ( -17.90, z-score = 0.84, R) >triCas2.chrUn_44 211943 99 + 525920 ---------ACUCAUCAAGAACGGGAAACUCAUCAUUGAAAAAACGAAAGACGACGAUGAGAAACAACUGAUCCAAUCCACUAUCAACAACUUGAGUGAUCAGCUGCA------ ---------.((((((..((..((....))..))..........(....).....))))))...((.((((((..........((((....))))..)))))).))..------ ( -19.80, z-score = -1.84, R) >consensus _________GGUCAAGACCAACGGCCAGCUGAUCAUCGAGAACUGUCGCAACAGCGAGGAGAAGACGCUGGUGCAGACGACCAUCGAUCAGUUGGCCGCAUCGCUGGC______ .....................(((((((......(((((........(((.((((...........)))).))).........)))))...)))))))................ (-15.01 = -14.24 + -0.77)


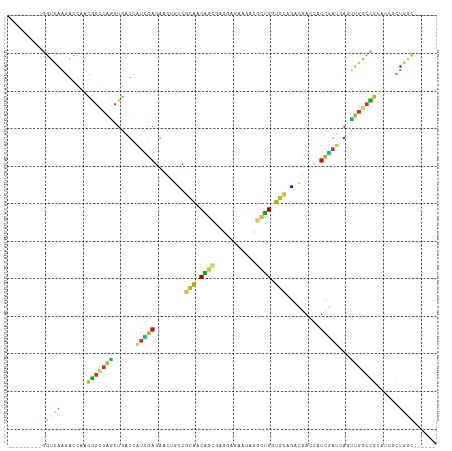
| Location | 5,154,775 – 5,154,874 |
|---|---|
| Length | 99 |
| Sequences | 15 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 73.63 |
| Shannon entropy | 0.57206 |
| G+C content | 0.55074 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -12.41 |
| Energy contribution | -12.00 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 2.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.880039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5154775 99 - 23011544 ------GCCAGCGAUGCGGCCAACUGAUCGAUGGUCGUCUGCACCAGCGUCUUCUCCUCGCUGUUGCGACAGUUCUCGAUGAUCAGCUGACCGUUGGUCUUGACC--------- ------(((((((...((((....(.(((((..(..(((.(((.(((((.........))))).))))))..)..))))).)...))))..))))))).......--------- ( -36.00, z-score = -1.39, R) >droSim1.chr2L 4962333 99 - 22036055 ------GCCAGCGAUGCGGCCAACUGAUCGAUGGUCGUCUGCACCAGCGUCUUCUCCUCGCUGUUGCGACAGUUCUCGAUGAUCAGCUGGCCGUUGGUCUUGACC--------- ------((((.....(((((((.((((((((..(..(((.(((.(((((.........))))).))))))..)..))))...)))).))))))))))).......--------- ( -40.00, z-score = -2.22, R) >droSec1.super_5 3241209 99 - 5866729 ------GCCAGCGAUGCGGCCAACUGAUCGAUGGUCGUCUGCACCAGCGUCUUCUCCUCGCUGUUGCGACAGUUCUCGAUGAUCAGCUGGCCGUUGGUCUUGACC--------- ------((((.....(((((((.((((((((..(..(((.(((.(((((.........))))).))))))..)..))))...)))).))))))))))).......--------- ( -40.00, z-score = -2.22, R) >droYak2.chr2L 11954959 99 + 22324452 ------GCCAGCGAUGCGGCCAACUGAUCGAUGGUCGUCUGCACCAGCGUCUUCUCCUCGCUGUUGCGACAGUUCUCGAUGAUCAGCUGGCCGUUGGUCUUGACC--------- ------((((.....(((((((.((((((((..(..(((.(((.(((((.........))))).))))))..)..))))...)))).))))))))))).......--------- ( -40.00, z-score = -2.22, R) >droEre2.scaffold_4929 5235922 99 - 26641161 ------GCCAGCGAUGCGGCCAACUGGUCGAUGGUCGUCUGCACCAGCGUCUUCUCCUCGCUGUUGCGACAGUUCUCGAUGAUCAGUUGGCCAUUGGUCUUGACC--------- ------((((((...))((((((((((((((..(..(((.(((.(((((.........))))).))))))..)..)))))...)))))))))..)))).......--------- ( -41.30, z-score = -2.73, R) >droAna3.scaffold_12916 6174614 99 + 16180835 ------GCCAGAGAGGCGGCCAACUGAUCGAUGGUUGUCUGCACCAGCGUCUUCUCCUCGCUGUUGCGACAGUUCUCGAUGAUCAGCUGGCCGUUGGUCUUAACC--------- ------...(((..((((((((.(((((((.((((((((.(((.(((((.........))))).))))))))...))).))))))).))))))))..))).....--------- ( -43.70, z-score = -3.56, R) >dp4.chr4_group3 8948911 99 + 11692001 ------GCCAGGGAAGCGGCCAACUGAUCGAUGGUCGUCUGCACCAGUGUCUUCUCCUCACUGUUGCGGCAAUUCUCGAUAAUCAGCUGGCCGUUGGUCUUGACC--------- ------..(((((.((((((((.((((((((..((.(.(((((.(((((.........))))).)))))).))..))))...)))).))))))))..)))))...--------- ( -42.60, z-score = -3.82, R) >droPer1.super_1 6054476 99 + 10282868 ------GCCAGGGAAGCGGCCAACUGAUCGAUGGUCGUCUGCACCAGUGUCUUCUCCUCACUGUUGCGGCAAUUCUCGAUAAUCAGCUGGCCGUUGGUCUUGACC--------- ------..(((((.((((((((.((((((((..((.(.(((((.(((((.........))))).)))))).))..))))...)))).))))))))..)))))...--------- ( -42.60, z-score = -3.82, R) >droWil1.scaffold_180772 7782175 99 + 8906247 ------GCCAAUGAGGCAGCCAAUUGAUCGAUUGUCGUCUGCACCAGUGUCUUCUCCUCACUGUUGCGACAAUUCUCAAUAAUUAGCUGUCCAUUUGUUUUGACC--------- ------((.((((.((((((..(((((..(((((((....(((.(((((.........))))).)))))))))).))))).....)))))))))).)).......--------- ( -29.30, z-score = -3.43, R) >droVir3.scaffold_12963 13764433 99 + 20206255 ------GCUAGCUGCGCGGCCAGCUGAUCAAUGGUCGAUUGCACCAGCGUCUUCUCCUCGCUGUUGCGACAGUUCUCAAUGAUCAGCUGACCGUUGGUCUUGACC--------- ------((.....))((((.((((((((((.(((.(..(((((.(((((.........))))).)))))..)...))).)))))))))).)))).(((....)))--------- ( -40.70, z-score = -3.20, R) >droMoj3.scaffold_6500 6765078 99 + 32352404 ------GCUAGCUGAGCGGCCAACUGAUCAAUGGUCGACUGCACCAGCGUCUUCUCCUCGCUGUUGCGACAAUUCUCAAUGAUCAGCUGGCCGUUGGUCUUGACC--------- ------....(((.((((((((.(((((((.(((.....((((.(((((.........))))).)))).......))).))))))).))))))))))).......--------- ( -38.10, z-score = -3.07, R) >droGri2.scaffold_15252 7522010 99 + 17193109 ------GCCAGCUGGGCGGCCAACUGAUCAAUGGUCGAUUGCACCAGCGUCUUCUCCUCGCUGUUGCGACAGUUCUCAAUGAUCAGCUGGCCGUUGGUCUUGACC--------- ------((((.....(((((((.(((((((.(((.(..(((((.(((((.........))))).)))))..)...))).))))))).))))))))))).......--------- ( -41.20, z-score = -3.02, R) >anoGam1.chr3R 16620053 114 + 53272125 UACAGCCGCAUAAAGUUCGCCCAAUUGCCGAGGAUCUGAUCGACCGUCUGGUUCUUCUGCUCGAUCAGCGCGAUCGUUUUGUCCAGCUCGUGCGUAAGCUGAUCCAUGUUGGCC ....(((((((.....((((......).)))((((((((((((..((..((....)).)))))))))((((((.(..........).)))))).......)))))))).)))). ( -30.80, z-score = 0.47, R) >apiMel3.Group1 9427581 99 + 25854376 ------UUCAACUGUUCGGUCACGUUGUGAACGGUGCUGUCGAUCAAUUGUUUCUCCGUGUCGUCCCUCGUGUUCUCCGCAAUCGUACGGCCAUUUCUCGUUAAU--------- ------....((.((((((.((((..(.((.(((..(.(..((.........))..))..))))))..))))....))).))).))((((.......))))....--------- ( -16.20, z-score = 1.39, R) >triCas2.chrUn_44 211943 99 - 525920 ------UGCAGCUGAUCACUCAAGUUGUUGAUAGUGGAUUGGAUCAGUUGUUUCUCAUCGUCGUCUUUCGUUUUUUCAAUGAUGAGUUUCCCGUUCUUGAUGAGU--------- ------.((((((((((...(((.(..(.....)..).)))))))))))))..(((((((.((...((((((........)))))).....))....))))))).--------- ( -24.20, z-score = -0.88, R) >consensus ______GCCAGCGAUGCGGCCAACUGAUCGAUGGUCGUCUGCACCAGCGUCUUCUCCUCGCUGUUGCGACAGUUCUCGAUGAUCAGCUGGCCGUUGGUCUUGACC_________ .................((.((.(((((....((.....((((.(((((.........))))).)))).....))......))))).)).))...................... (-12.41 = -12.00 + -0.41)

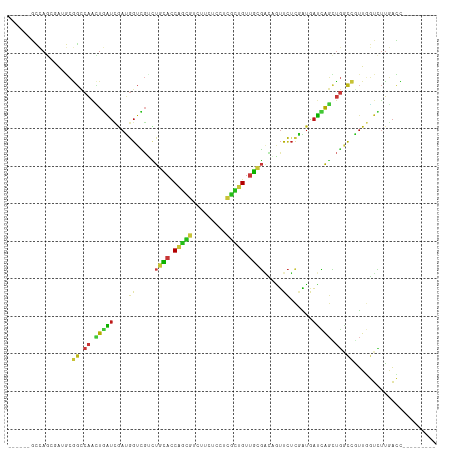
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:13 2011