| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,668,913 – 13,669,009 |
| Length | 96 |
| Max. P | 0.888596 |

| Location | 13,668,913 – 13,669,009 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 69.51 |
| Shannon entropy | 0.63998 |
| G+C content | 0.44115 |
| Mean single sequence MFE | -18.40 |
| Consensus MFE | -11.39 |
| Energy contribution | -11.64 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
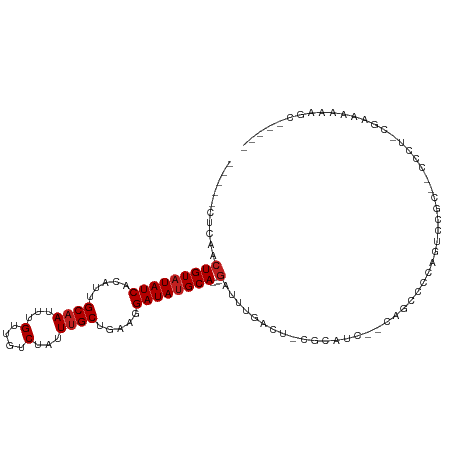
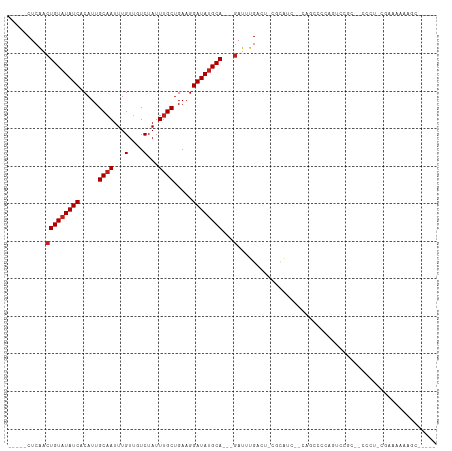
>dm3.chr3L 13668913 96 - 24543557 -----CUCAACUGUAUAUCACAUUGCAAUUUGUUGUCUAUUUGCUGAAGGAUAUGCA---GAUUUGACU-CUCAUG--CAGCCCCAGUCCGC--CCCUCCGAAAAAAGC----- -----.((..(((((((((.....((((...(....)...)))).....))))))))---)....((((-......--.......))))...--......)).......----- ( -16.52, z-score = 0.21, R) >droSim1.chr3L 13054184 96 - 22553184 -----CUCAACUGUAUAUCACAUUGCAAUUUGUUGUCUAUUUGCUGAAGGAUAUGCA---GAUUUGACU-CUCAUC--CAGCCCCAGUCCGC--CCCUCCAAAAAAAGC----- -----.....(((((((((.....((((...(....)...)))).....))))))))---)....((((-......--.......))))...--...............----- ( -16.32, z-score = -0.50, R) >droSec1.super_0 5831119 96 - 21120651 -----CUCAACUGUAUAUCACAUUGCAAUUUGUUGUCUAUUUGCUGAAGGAUAUGCA---GAUUUGACU-CUCAUC--CAGCCCCAGUCCGC--CCCUCCGAAAAAAGC----- -----.((..(((((((((.....((((...(....)...)))).....))))))))---)....((((-......--.......))))...--......)).......----- ( -16.52, z-score = -0.32, R) >droYak2.chr3L 13764398 100 - 24197627 -----CUCAACUGUAUAUCACAUUGCAAUUUGUUGUCUAUUUGCUGAAGGAUAUGCA---GAUUUGACU-CGCAUCCACAGUUCCACUCCGCCGCCCCUCGAAAAAAGC----- -----.....(((((((((.....((((...(....)...)))).....))))))))---).(((((..-.((......((.....)).....))...)))))......----- ( -17.40, z-score = -0.07, R) >droEre2.scaffold_4784 13671912 95 - 25762168 -----CUCAACUGUAUAUCACAUUGCAAUUUGUUGUCUAUUUGCUGAAGGAUAUGCA---GAUUUGACU-CGCAUC--CAGCCCCAGUCCGC--CCCU-CGAAAAAAGC----- -----.((..(((((((((.....((((...(....)...)))).....))))))))---)....((((-.((...--..))...))))...--....-.)).......----- ( -19.10, z-score = -0.85, R) >droAna3.scaffold_13337 1523212 79 + 23293914 -----CUCGUCUGUAUAUCACAUUGCCAUUUGUUGUCUAUUUGCUGAAGGAUACGCA---GAUUUGACUCGUCGCCUCGAAAAAAUA--------------------------- -----...(((((..((((.....((.....(....).....)).....))))..))---))).....(((......))).......--------------------------- ( -12.10, z-score = 0.65, R) >dp4.chrXR_group8 2454434 81 + 9212921 -----CUCAACUGUAUAUCACAUUGCAAUUUGUUGUCUAUUUGCUGAAGGAUAUGCA---GAUGUGACUCCCUGCCUCGACAAAAAAAG------------------------- -----(.((.(((((((((.....((((...(....)...)))).....))))))))---).)).).......................------------------------- ( -16.30, z-score = -1.25, R) >droPer1.super_36 549283 81 - 818889 -----CUCAACUGUAUAUCACAUUGCAAUUUGUUGUCUAUUUGCUGAAGGAUAUGCA---GAUGUGACUCCCUGCCUCGACAAAAAAAG------------------------- -----(.((.(((((((((.....((((...(....)...)))).....))))))))---).)).).......................------------------------- ( -16.30, z-score = -1.25, R) >droWil1.scaffold_180698 10894303 106 + 11422946 -----CUCAACUGUAUAUCACAUUGCAAUUUGUUGUCUAUUUGCUGAAGGAUAUGCACAGAAUAAGAGAGAAAAAGGGCCAAAUCAGCCCAUCAGCCCAUGAGAUGGGCUA--- -----(((...((((((((.....((((...(....)...)))).....))))))))(.......).))).....((((.......))))...(((((((...))))))).--- ( -28.60, z-score = -1.32, R) >droMoj3.scaffold_6680 17326799 109 + 24764193 GGCAGCUCAACUGUAUAUCACAUUGCAAUUUGUUGUCUAUUUGCUGAAGGAUAUGCA---GAUUUGCCA-GCCGCCUCGCCCCACGACCCACGCCCCUCGUCCUGCCACGUCC- (((((..(..(((((((((.....((((...(....)...)))).....))))))))---)........-((....(((.....))).....)).....)..)))))......- ( -26.40, z-score = -1.07, R) >droVir3.scaffold_13049 5504424 106 + 25233164 -----CUCAACUGUAUAUCACAUUGCAAUUUGUUGUCUAUUUGCUGAAGGAUAUGCA---GAUUUGCCUGCGCCUCGAAUACCCUUUCCUUUUGCCCCUCUCCCCCCCACUUAC -----.....(((((((((.....((((...(....)...)))).....))))))))---)........(((....(((......)))....)))................... ( -17.10, z-score = -1.86, R) >droGri2.scaffold_15110 17626262 92 - 24565398 -----CUCAACUGUAUAUCACAUUGCAAUUUGUUGUCUAUUUGCUGAAGGAUAUGCA---GAUUUGCCUGCGCCUC-AAUG-CCUGCCUCACUGACACUCUU------------ -----.(((.(((((((((.....((((...(....)...)))).....))))))))---)....((..(((....-..))-)..)).....))).......------------ ( -18.20, z-score = -0.39, R) >consensus _____CUCAACUGUAUAUCACAUUGCAAUUUGUUGUCUAUUUGCUGAAGGAUAUGCA___GAUUUGACU_CGCAUC__CAGCCCCAGUCCGC__CCCU_CGAAAAAAGC_____ ......((...((((((((.....((((...(....)...)))).....))))))))...)).................................................... (-11.39 = -11.64 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:03 2011