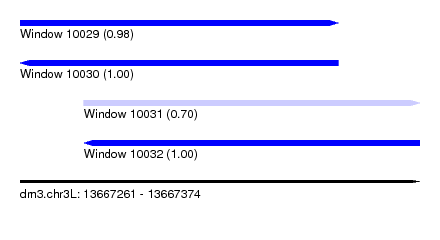
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,667,261 – 13,667,374 |
| Length | 113 |
| Max. P | 0.998743 |

| Location | 13,667,261 – 13,667,351 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 75.52 |
| Shannon entropy | 0.51609 |
| G+C content | 0.51531 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -14.15 |
| Energy contribution | -13.44 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
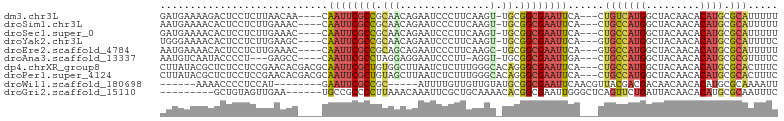
>dm3.chr3L 13667261 90 + 24543557 -GCCUGGAAAAGUGUGCGGAAAAAUGCGCAUGUGUUGUAGCCAUGACAGUGAAUUCGCCGCA-ACUUGAAGGGAUUCUGUUGCGGCGAAUUG- -(.(((.((..(((((((......)))))))...)).))).).........(((((((((((-((..(((....))).))))))))))))).- ( -35.50, z-score = -3.74, R) >droSim1.chr3L 13052520 90 + 22553184 -GCCUGGAAAAGUGUGCGGAAAAAUGCGCAUGUGUUGUAGCCAUGGCAGUGAAUUCGCCGCA-ACUUGAAGGGAUUCUGUUGCGGCGAAUUG- -((((((.((.(((((((......)))))))...))....))).)))....(((((((((((-((..(((....))).))))))))))))).- ( -40.60, z-score = -4.98, R) >droSec1.super_0 5829473 90 + 21120651 -GCCUGGAAAAGUGUGCGGAAAAAUGCGCAUGUGUUGUAGCCAUGGCAGUGAAUUCGCCGCA-ACUUGAAGGGAUUCUGUUGCGGCGAAUUG- -((((((.((.(((((((......)))))))...))....))).)))....(((((((((((-((..(((....))).))))))))))))).- ( -40.60, z-score = -4.98, R) >droYak2.chr3L 13762687 90 + 24197627 -GCCUGGAAAAGCGUGCGGGAAAAUGCGCAUGUGUUGUAGCCAUGGCACUGAAUUCGCCGCA-ACUUGAAGGGAUUCUGUUGCGGCGAAUUG- -((((((.((.(((((((........))))))).))....))).)))....(((((((((((-((..(((....))).))))))))))))).- ( -42.10, z-score = -4.70, R) >droEre2.scaffold_4784 13670254 90 + 25762168 -GCCUGGAAAAGUGUGCGGAAAAAUGCGCAUGUGUUGUAGCCAUGGCACUGAAUUCGCCGCA-GCUUGAAGGGAUUCUGCUGCGGCGAAUUG- -((((((.((.(((((((......)))))))...))....))).)))....(((((((((((-((..(((....))).))))))))))))).- ( -43.00, z-score = -4.99, R) >droAna3.scaffold_13337 1521609 89 - 23293914 GGCCAGCAAAGUGG-UCUGGAAAACGCGCAUGUGUUGUAGCCAUGGCAGUCAAUUCGCC--GCAACCUAAGGGAUUCCUCCUAGGCGAAUUG- (((..((...((((-.(((...(((((....))))).))))))).)).)))((((((((--.........((((....)))).)))))))).- ( -27.10, z-score = -0.34, R) >dp4.chrXR_group8 2452640 92 - 9212921 -UCCUGGAAGGCGGGCCUGGAAAGUGCGCAUGUGUUGUAGCCAUGGCAGUGAAUUCGCCCUGUGCCCAAAGAGAUUAAGCCACAGCGAAUUGC -(((((...((((((((.((....(((.((((.((....)))))))))(((....))))).).)))).((....))..))).))).))..... ( -26.10, z-score = 0.70, R) >droPer1.super_4124 173 92 + 4472 -UCCUGGAAGGCGGGCCUGGAAAGUGCGCAUGUGUUGUAGCCAUGGCAGUGAAUUCGCCCUGUGCCCAAAGAGAUUAAGCUACAGCGAAUUGC -(((((...((((((((.((....(((.((((.((....)))))))))(((....))))).).)))).((....))..))).))).))..... ( -24.20, z-score = 1.22, R) >droWil1.scaffold_180698 10890362 85 - 11422946 -UUCUGGAAAAUUGAAAUUUUGCGCAUGUGU-UGUUGUCGUCGUAACGUUGAAUUCGCCGCAUACAACAACAAAAU-----GCGGCGAAUUC- -..............(((.(((((.(((...-......)))))))).)))(((((((((((((...........))-----)))))))))))- ( -24.10, z-score = -1.45, R) >consensus _GCCUGGAAAAGUGUGCGGAAAAAUGCGCAUGUGUUGUAGCCAUGGCAGUGAAUUCGCCGCA_ACUUGAAGGGAUUCUGCUGCGGCGAAUUG_ ........................(((.((((.((....)))))))))...((((((((.((.((.............)))).)))))))).. (-14.15 = -13.44 + -0.71)

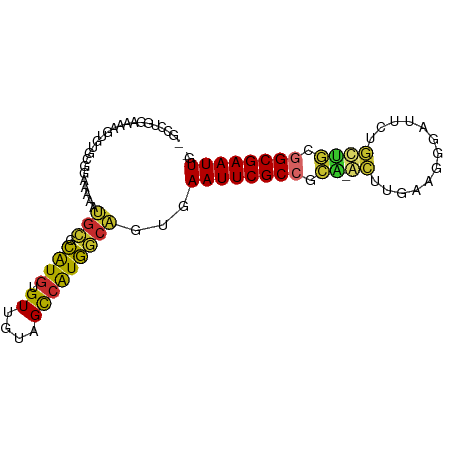
| Location | 13,667,261 – 13,667,351 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 75.52 |
| Shannon entropy | 0.51609 |
| G+C content | 0.51531 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -15.30 |
| Energy contribution | -14.09 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.63 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.996428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13667261 90 - 24543557 -CAAUUCGCCGCAACAGAAUCCCUUCAAGU-UGCGGCGAAUUCACUGUCAUGGCUACAACACAUGCGCAUUUUUCCGCACACUUUUCCAGGC- -.(((((((((((((.(((....)))..))-)))))))))))....(((.(((..........((((........)))).......))))))- ( -26.33, z-score = -3.51, R) >droSim1.chr3L 13052520 90 - 22553184 -CAAUUCGCCGCAACAGAAUCCCUUCAAGU-UGCGGCGAAUUCACUGCCAUGGCUACAACACAUGCGCAUUUUUCCGCACACUUUUCCAGGC- -.(((((((((((((.(((....)))..))-)))))))))))....(((.(((..........((((........)))).......))))))- ( -29.03, z-score = -4.35, R) >droSec1.super_0 5829473 90 - 21120651 -CAAUUCGCCGCAACAGAAUCCCUUCAAGU-UGCGGCGAAUUCACUGCCAUGGCUACAACACAUGCGCAUUUUUCCGCACACUUUUCCAGGC- -.(((((((((((((.(((....)))..))-)))))))))))....(((.(((..........((((........)))).......))))))- ( -29.03, z-score = -4.35, R) >droYak2.chr3L 13762687 90 - 24197627 -CAAUUCGCCGCAACAGAAUCCCUUCAAGU-UGCGGCGAAUUCAGUGCCAUGGCUACAACACAUGCGCAUUUUCCCGCACGCUUUUCCAGGC- -.(((((((((((((.(((....)))..))-))))))))))).(((((((((.........)))).))))).........(((......)))- ( -30.10, z-score = -3.45, R) >droEre2.scaffold_4784 13670254 90 - 25762168 -CAAUUCGCCGCAGCAGAAUCCCUUCAAGC-UGCGGCGAAUUCAGUGCCAUGGCUACAACACAUGCGCAUUUUUCCGCACACUUUUCCAGGC- -.(((((((((((((.(((....)))..))-))))))))))).(((((((((.........)))).))))).....................- ( -31.50, z-score = -3.91, R) >droAna3.scaffold_13337 1521609 89 + 23293914 -CAAUUCGCCUAGGAGGAAUCCCUUAGGUUGC--GGCGAAUUGACUGCCAUGGCUACAACACAUGCGCGUUUUCCAGA-CCACUUUGCUGGCC -(((((((((...((((....)))).......--)))))))))...((((((.........)))).)).....((((.-(......))))).. ( -25.00, z-score = -0.43, R) >dp4.chrXR_group8 2452640 92 + 9212921 GCAAUUCGCUGUGGCUUAAUCUCUUUGGGCACAGGGCGAAUUCACUGCCAUGGCUACAACACAUGCGCACUUUCCAGGCCCGCCUUCCAGGA- (.((((((((.((((((((.....)))))).)).)))))))))..(((((((.........)))).)))...((((((....)))....)))- ( -28.80, z-score = -0.89, R) >droPer1.super_4124 173 92 - 4472 GCAAUUCGCUGUAGCUUAAUCUCUUUGGGCACAGGGCGAAUUCACUGCCAUGGCUACAACACAUGCGCACUUUCCAGGCCCGCCUUCCAGGA- (.((((((((.(.((((((.....))))))..).)))))))))..(((((((.........)))).)))...((((((....)))....)))- ( -24.20, z-score = 0.17, R) >droWil1.scaffold_180698 10890362 85 + 11422946 -GAAUUCGCCGC-----AUUUUGUUGUUGUAUGCGGCGAAUUCAACGUUACGACGACAACA-ACACAUGCGCAAAAUUUCAAUUUUCCAGAA- -(((((((((((-----((...........)))))))))))))..(((....)))......-..............................- ( -22.80, z-score = -1.93, R) >consensus _CAAUUCGCCGCAACAGAAUCCCUUCAAGU_UGCGGCGAAUUCACUGCCAUGGCUACAACACAUGCGCAUUUUCCCGCACACUUUUCCAGGC_ ..((((((((.((((.............)).)).))))))))...(((((((.........)))).)))........................ (-15.30 = -14.09 + -1.22)

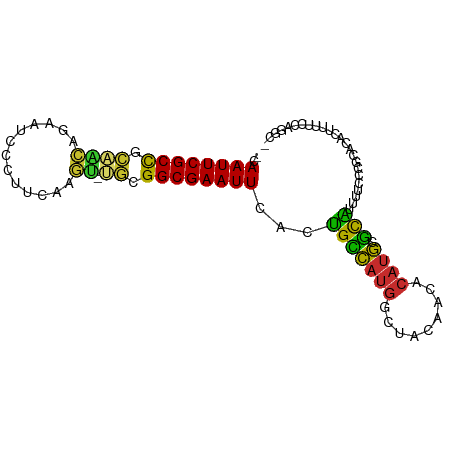
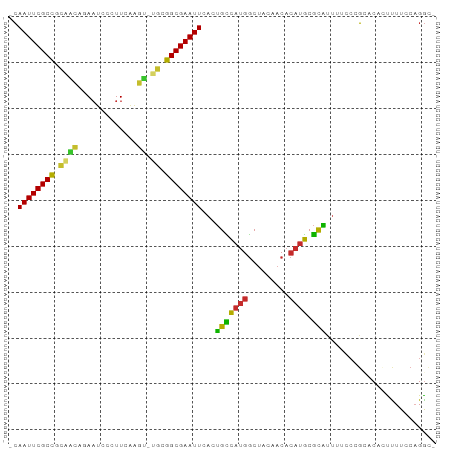
| Location | 13,667,279 – 13,667,374 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 68.43 |
| Shannon entropy | 0.67569 |
| G+C content | 0.47792 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -10.34 |
| Energy contribution | -11.12 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.698409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13667279 95 + 24543557 AAAAAUGCGCAUGUGUUGUAGCCAUGACAG---UGAAUUCGCCGCA-ACUUGAAGGGAUUCUGUUGCGGCGAAUUG----UUGUUAAGAGGAGUCUUUUCAUC ......(((((.....))).))..((((((---..(.(((((((((-((..(((....))).))))))))))))..----)))))).(((((...)))))... ( -31.00, z-score = -2.55, R) >droSim1.chr3L 13052538 95 + 22553184 AAAAAUGCGCAUGUGUUGUAGCCAUGGCAG---UGAAUUCGCCGCA-ACUUGAAGGGAUUCUGUUGCGGCGAAUUG----GUUUCAAGAGGAGUGUUUUCAUU ......((((.(.(.(((.(((((......---...((((((((((-((..(((....))).))))))))))))))----))).))).).).))))....... ( -32.60, z-score = -2.53, R) >droSec1.super_0 5829491 95 + 21120651 AAAAAUGCGCAUGUGUUGUAGCCAUGGCAG---UGAAUUCGCCGCA-ACUUGAAGGGAUUCUGUUGCGGCGAAUUG----GUUUCAAGAGGAGUGUUUUCAUC ......((((.(.(.(((.(((((......---...((((((((((-((..(((....))).))))))))))))))----))).))).).).))))....... ( -32.60, z-score = -2.59, R) >droYak2.chr3L 13762705 95 + 24197627 GAAAAUGCGCAUGUGUUGUAGCCAUGGCAC---UGAAUUCGCCGCA-ACUUGAAGGGAUUCUGUUGCGGCGAAUUG----GCUUCAAGAGGAGUGUUUUCCCA ((((((((...(.(.(((.(((((......---...((((((((((-((..(((....))).))))))))))))))----))).))).).).))))))))... ( -37.10, z-score = -2.97, R) >droEre2.scaffold_4784 13670272 95 + 25762168 AAAAAUGCGCAUGUGUUGUAGCCAUGGCAC---UGAAUUCGCCGCA-GCUUGAAGGGAUUCUGCUGCGGCGAAUUG----GUUUCAAGAGGAGUGUUUUCAUU .(((((((.((.((((..(....)..))))---))(((((((((((-((..(((....))).))))))))))))).----............))))))).... ( -35.20, z-score = -2.53, R) >droAna3.scaffold_13337 1521627 91 - 23293914 GAAAACGCGCAUGUGUUGUAGCCAUGGCAG---UCAAUUCGCCGCA-ACCU-AAGGGAUUCCUCCUAGGCGAAUUG----GGCUC---AGGGGUAUUGACAUU ...(((((....)))))(((.((.((((..---.(((((((((...-....-..((((....)))).)))))))))----.)).)---).)).)))....... ( -26.00, z-score = 0.07, R) >dp4.chrXR_group8 2452658 100 - 9212921 GAAAGUGCGCAUGUGUUGUAGCCAUGGCAG---UGAAUUCGCCCUGUGCCCAAAGAGAUUAAGCCACAGCGAAUUGCGUCGUGUUCGGAGGAGAGCGUAUAAG ....((((((............((((((.(---(.(((((((..((.((.............))))..))))))))))))))).((......))))))))... ( -25.02, z-score = 0.23, R) >droPer1.super_4124 191 100 + 4472 GAAAGUGCGCAUGUGUUGUAGCCAUGGCAG---UGAAUUCGCCCUGUGCCCAAAGAGAUUAAGCUACAGCGAAUUGCGUCGUGUUCGGAGGAGAGCGUAUAAG ....(((((((..(((((((((...(((((---((....)))....))))(.....).....)))))))))...))).....((((......))))))))... ( -27.60, z-score = -0.56, R) >droWil1.scaffold_180698 10890376 84 - 11422946 AAUUUUGCGCAUGUGUUGUUGUCGUCGUAACGUUGAAUUCGCCGCAUACAACAACAAAAU-----GCGGCGAAUUC--------AUGGAGGGGUUUU------ ....(((((.(((.........))))))))(((.(((((((((((((...........))-----)))))))))))--------)))..........------ ( -26.10, z-score = -1.50, R) >droGri2.scaffold_15110 17623187 88 + 24565398 GAAAUUGCGCAUGUGUUGUAAUCAGAACUGAGCCCAAUUCGCCGUGUUUUGCAGCGAAUUUGUUUAAGGGGCGGCA------UUCAACUACAGC--------- ........((.(((((((...(((....)))(((((((((((.((.....)).)))))))........))))....------..)))).)))))--------- ( -21.60, z-score = 0.36, R) >consensus AAAAAUGCGCAUGUGUUGUAGCCAUGGCAG___UGAAUUCGCCGCA_ACUUGAAGGGAUUCUGUUGCGGCGAAUUG____GUUUCAAGAGGAGUGUUUUCAUU .....(((.((((.((....)))))))))......((((((((........................))))))))............................ (-10.34 = -11.12 + 0.78)
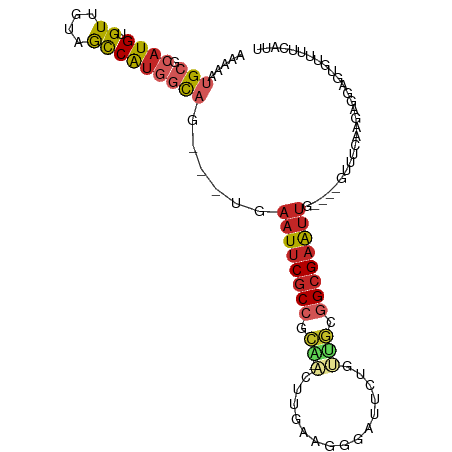
| Location | 13,667,279 – 13,667,374 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 68.43 |
| Shannon entropy | 0.67569 |
| G+C content | 0.47792 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -10.40 |
| Energy contribution | -10.61 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.998743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
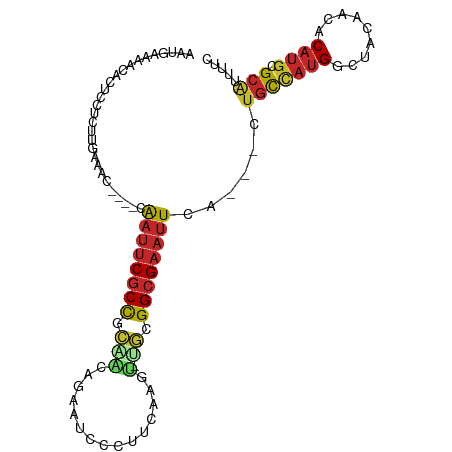
>dm3.chr3L 13667279 95 - 24543557 GAUGAAAAGACUCCUCUUAACAA----CAAUUCGCCGCAACAGAAUCCCUUCAAGU-UGCGGCGAAUUCA---CUGUCAUGGCUACAACACAUGCGCAUUUUU ..((((((((....)))).....----...(((((((((((.(((....)))..))-)))))))))))))---.(((((((.........)))).)))..... ( -25.40, z-score = -3.50, R) >droSim1.chr3L 13052538 95 - 22553184 AAUGAAAACACUCCUCUUGAAAC----CAAUUCGCCGCAACAGAAUCCCUUCAAGU-UGCGGCGAAUUCA---CUGCCAUGGCUACAACACAUGCGCAUUUUU .......................----.(((((((((((((.(((....)))..))-)))))))))))..---.(((((((.........)))).)))..... ( -27.40, z-score = -4.35, R) >droSec1.super_0 5829491 95 - 21120651 GAUGAAAACACUCCUCUUGAAAC----CAAUUCGCCGCAACAGAAUCCCUUCAAGU-UGCGGCGAAUUCA---CUGCCAUGGCUACAACACAUGCGCAUUUUU .......................----.(((((((((((((.(((....)))..))-)))))))))))..---.(((((((.........)))).)))..... ( -27.40, z-score = -4.23, R) >droYak2.chr3L 13762705 95 - 24197627 UGGGAAAACACUCCUCUUGAAGC----CAAUUCGCCGCAACAGAAUCCCUUCAAGU-UGCGGCGAAUUCA---GUGCCAUGGCUACAACACAUGCGCAUUUUC .((((......))))........----.(((((((((((((.(((....)))..))-))))))))))).(---((((((((.........)))).)))))... ( -31.80, z-score = -3.55, R) >droEre2.scaffold_4784 13670272 95 - 25762168 AAUGAAAACACUCCUCUUGAAAC----CAAUUCGCCGCAGCAGAAUCCCUUCAAGC-UGCGGCGAAUUCA---GUGCCAUGGCUACAACACAUGCGCAUUUUU .......................----.(((((((((((((.(((....)))..))-))))))))))).(---((((((((.........)))).)))))... ( -31.50, z-score = -4.41, R) >droAna3.scaffold_13337 1521627 91 + 23293914 AAUGUCAAUACCCCU---GAGCC----CAAUUCGCCUAGGAGGAAUCCCUU-AGGU-UGCGGCGAAUUGA---CUGCCAUGGCUACAACACAUGCGCGUUUUC ...((((((......---..(((----((....(((((((.((...)))))-))))-)).)))..)))))---).((((((.........)))).))...... ( -23.40, z-score = -0.35, R) >dp4.chrXR_group8 2452658 100 + 9212921 CUUAUACGCUCUCCUCCGAACACGACGCAAUUCGCUGUGGCUUAAUCUCUUUGGGCACAGGGCGAAUUCA---CUGCCAUGGCUACAACACAUGCGCACUUUC .......((((............)).))((((((((.((((((((.....)))))).)).))))))))..---.(((((((.........)))).)))..... ( -25.70, z-score = -2.05, R) >droPer1.super_4124 191 100 - 4472 CUUAUACGCUCUCCUCCGAACACGACGCAAUUCGCUGUAGCUUAAUCUCUUUGGGCACAGGGCGAAUUCA---CUGCCAUGGCUACAACACAUGCGCACUUUC ......((((((.......((((((......))).))).((((((.....))))))..))))))......---.(((((((.........)))).)))..... ( -22.90, z-score = -1.34, R) >droWil1.scaffold_180698 10890376 84 + 11422946 ------AAAACCCCUCCAU--------GAAUUCGCCGC-----AUUUUGUUGUUGUAUGCGGCGAAUUCAACGUUACGACGACAACAACACAUGCGCAAAAUU ------............(--------(((((((((((-----((...........)))))))))))))).(((....)))...................... ( -23.80, z-score = -2.82, R) >droGri2.scaffold_15110 17623187 88 - 24565398 ---------GCUGUAGUUGAA------UGCCGCCCCUUAAACAAAUUCGCUGCAAAACACGGCGAAUUGGGCUCAGUUCUGAUUACAACACAUGCGCAAUUUC ---------(.((((((((((------(...((((........(((((((((.......)))))))))))))...)))).))))))).).............. ( -24.80, z-score = -2.13, R) >consensus AAUGAAAACACUCCUCUUGAAAC____CAAUUCGCCGCAACAGAAUCCCUUCAAGU_UGCGGCGAAUUCA___CUGCCAUGGCUACAACACAUGCGCAUUUUC ............................((((((((.((..................)).))))))))......(((((((.........)))).)))..... (-10.40 = -10.61 + 0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:24:02 2011