
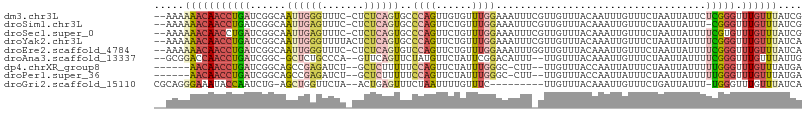
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,659,218 – 13,659,323 |
| Length | 105 |
| Max. P | 0.787400 |

| Location | 13,659,218 – 13,659,323 |
|---|---|
| Length | 105 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.10 |
| Shannon entropy | 0.46478 |
| G+C content | 0.35517 |
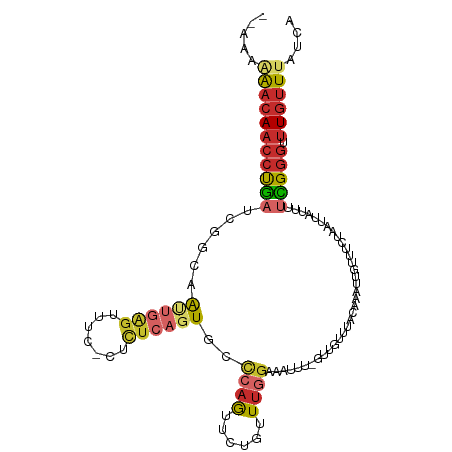
| Mean single sequence MFE | -22.59 |
| Consensus MFE | -10.16 |
| Energy contribution | -11.32 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.787400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13659218 105 + 24543557 --AAAAAACAACCUGAUCGGCAAUUGGGUUUC-CUCUCAGUGCCCAGUUGUGUUUGGAAAUUUCGUUGUUUACAAUUUGUUUCUAAUUAUUCUCGGGUUUGUUUAUCG --...(((((((((((...((((((((((...-........))))))))))..((((((((...((((....))))..))))))))......))))).)))))).... ( -31.80, z-score = -4.78, R) >droSim1.chr3L 13044418 104 + 22553184 --AAAAAACAACCUGAUCGGCAAUUGAGUUUC-CUCUCAGUGCCCAGUUCUGUUUGGAAAUUUCGUUGUUUACAAAUUGUUUCUAAUUAUUU-CGGGUUUGUUUAUCG --...(((((((((((..((((.(((((....-..))))))))).........((((((((....(((....)))...)))))))).....)-)))).)))))).... ( -28.50, z-score = -4.15, R) >droSec1.super_0 5821540 105 + 21120651 --AAAAAACAACCUGAUCGGCAAUUGAGUUUC-CUCUCAGUGCCCAGUUCUGUUUGGAAAUUUCGUUGUUUACAAAUUGUUUCUAAUUAUUUUCGUGUUUGUUUAUCG --...(((((((.(((..((((.(((((....-..))))))))).........((((((((....(((....)))...))))))))......))).).)))))).... ( -22.00, z-score = -2.60, R) >droYak2.chr3L 13754037 106 + 24197627 --AAAAAACAACCUGAUCGGCAAUUGGGUUUUACUCUCAGUGCCCAGUUCUGUUUGGAAAUUUCGUUGUUUACAAAUUGUUUCUAAUUAUUUUCGGGUUUGUUUAUCA --...(((((((((((.(((.((((((((............))))))))))).((((((((....(((....)))...))))))))......))))).)))))).... ( -28.70, z-score = -3.75, R) >droEre2.scaffold_4784 13662018 105 + 25762168 --AAAAAACAACCUGAUCGGCAAUUGGGUUUC-CUCUCAGUGUCCAGUUCUGUUUGGAAAUUUGGUUGUUUACAAAUUGUUUCUAAUUAUUUUCGGGUUUGUUUAUCA --...(((((((((((..((((((..((((((-(...(((.........)))...)))))))..))))))....(((((....)))))....))))).)))))).... ( -28.20, z-score = -3.48, R) >droAna3.scaffold_13337 1511686 101 - 23293914 --GCGGACCAACCUGAUCGGC-GCUCUGCCCA--GUUCAGUUCUAUGUUCUAUUCGGACAUUU--UUGUUUACAAAUUGUUUCUAAUUAUUUUCGGGUUUGUUUAUUG --..((((.(((((((..(((-.....)))..--((.(((....((((((.....))))))..--)))...))...................))))))).)))).... ( -20.50, z-score = -1.16, R) >dp4.chrXR_group8 2444705 97 - 9212921 ------AACAACCUGAUCGGCAGCCGAGAUCU--GCUCUUUUUCCAGUUCUAUUUGGGC-CUU--UUGUUUACCAAUUAUUUCUAAUUAUUUUUGGGUUUGUUUAUGA ------(((((((..(..(((....(((....--.))).....((((......))))))-)..--.........(((((....)))))....)..)).)))))..... ( -16.10, z-score = 0.17, R) >droPer1.super_36 539002 97 + 818889 ------AACAACCUGAUCGGCAGCCGAGAUCU--GCUCUUUUUCCAGUUCUAUUUGGGC-CUU--UUGUUUACCAAUUAUUUCUAAUUAUUUUUGGGUUUGUUUAUGA ------(((((((..(..(((....(((....--.))).....((((......))))))-)..--.........(((((....)))))....)..)).)))))..... ( -16.10, z-score = 0.17, R) >droGri2.scaffold_15110 17613267 95 + 24565398 CGCAGGGAAAUACCAAUCUG-AGCUGGUUCUA--ACUGAGUUUCUAAUUUUGUUUC---------UUGUUUACAAAUUGUUUCUGAUUAUUU-UGGGUUUGUUUAUCA .((((((((..........(-(((((((....--))).))))).........))))---------))))..((((((.....(..(.....)-..)))))))...... ( -11.41, z-score = 1.58, R) >consensus __AAAAAACAACCUGAUCGGCAAUUGAGUUUC_CUCUCAGUGCCCAGUUCUGUUUGGAAAUUU_GUUGUUUACAAAUUGUUUCUAAUUAUUUUCGGGUUUGUUUAUCA .....(((((((((((..((..((((((.......))))))..))........(((((.......(((....)))......)))))......))))).)))))).... (-10.16 = -11.32 + 1.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:59 2011