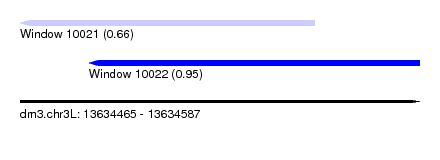
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,634,465 – 13,634,587 |
| Length | 122 |
| Max. P | 0.952069 |

| Location | 13,634,465 – 13,634,555 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 84.90 |
| Shannon entropy | 0.28704 |
| G+C content | 0.54731 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -16.85 |
| Energy contribution | -16.85 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13634465 90 - 24543557 UUAGCGCUUGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAUCAUAGCCCC--CCGCCCAAAAAGGACCACCCACAACAACGC ...(((.((((((...(((((((.....)))))))((((......))))...........--..))))))...((.....)).......))) ( -23.10, z-score = -1.85, R) >droSim1.chr3L 13018279 86 - 22553184 UUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAGCACAGCCCC--CCG----AAAAGGACCACCCACAACAACGC ...(((.(((((....(((((((.....)))))))((((...(((.....)))..)))))--)))----)...((.....)).......))) ( -22.60, z-score = -1.69, R) >droSec1.super_0 5796281 87 - 21120651 UUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAGCACAGCCCC--CC---CAAAAAGGACCACCCACAACAACGC ...(((...((((.(((((((((.....))))))))).....(((.....)))..)))).--..---......((.....)).......))) ( -20.90, z-score = -1.37, R) >droYak2.chr3L 13728792 92 - 24197627 UUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCAGCCAGCAAAGCCCCAACUCCCCAAAAAGGACCUCCCACAACAACGC ...(((...((((.(((((((((.....))))))))).....(((.....)))..))))..............((.....)).......))) ( -21.20, z-score = -1.03, R) >droEre2.scaffold_4784 13637186 91 - 25762168 UUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAGCACAGCCCCCGCCCCCCAAAAAGGACCUCCCACAACAGCG- ....((((.((((...(((((((.....)))))))((((...(((.....)))..))))..))))........((.....)).....))))- ( -24.30, z-score = -1.96, R) >droAna3.scaffold_13337 1487362 78 + 23293914 UUAGCGCUCGGGCAUCAUAAACUAACGGAGUUUAUGGGUCCAUGCACCCAGGACACUCC----------AUGCUGACUG-CCACAUCGU--- (((((((..((((.(((((((((.....)))))))))))))..)).....((.....))----------..)))))...-.........--- ( -23.90, z-score = -1.66, R) >consensus UUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAGCACAGCCCC__CC___CAAAAAGGACCACCCACAACAACGC .....((..((((.(((((((((.....)))))))))))))..))............................................... (-16.85 = -16.85 + -0.00)

| Location | 13,634,486 – 13,634,587 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.00 |
| Shannon entropy | 0.35510 |
| G+C content | 0.54562 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -21.67 |
| Energy contribution | -21.76 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13634486 101 - 24543557 ----------CCCUGUCGCUCUGCGUAUUUCGCUUUGCUCACUUAGCGCUUGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAUCAUAGCCCC--CCGCCCAAA ----------.......((...(((.....)))...)).......(((...((((...(((((((.....)))))))((((......))))......)))).--.)))..... ( -26.70, z-score = -1.57, R) >droSim1.chr3L 13018300 97 - 22553184 ----------CCCUGUCGCUCUGCGUAUUUCGCUUUGCUCACUUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAGCACAGCCCC-----CCGAA- ----------..((((.(((.(((((....((((..(....)..))))...((((.(((((((((.....))))))))))))))))))...)))))))....-----.....- ( -29.00, z-score = -1.91, R) >droSec1.super_0 5796302 98 - 21120651 ----------CCCUGUCGCUCUGCGUAUUUCGCUUUGCUCACUUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAGCACAGCCCC-----CCCAAA ----------..((((.(((.(((((....((((..(....)..))))...((((.(((((((((.....))))))))))))))))))...)))))))....-----...... ( -29.00, z-score = -2.44, R) >droYak2.chr3L 13728813 103 - 24197627 ----------CCCUGUCGCUCUGCGUAUUUCGCUUUGCUCACUUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCAGCCAGCAAAGCCCCAACUCCCCAAA ----------.......(((((((((....((((..(....)..))))...((((.(((((((((.....)))))))))))))))))))..)))................... ( -28.50, z-score = -1.67, R) >droEre2.scaffold_4784 13637206 103 - 25762168 ----------CCCCGUCGCUCUGCGUAUUUCGCUUUGCUCACUUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAGCACAGCCCCCGCCCCCCAAA ----------.......(((.(((.......(((..(....)..)))((..((((.(((((((((.....)))))))))))))..)).....))).))).............. ( -27.40, z-score = -1.46, R) >droAna3.scaffold_13337 1487378 99 + 23293914 ----CCCGAUUGCUGUC-CUCUGCGAUUUUCGCUUUGCUCACUUAGCGCUCGGGCAUCAUAAACUAACGGAGUUUAUGGGUCCAUGCACCCAGGACACUCCAUG--------- ----(((((.(((((..-....((((........)))).....))))).)))))..............(((((...(((((......)))))....)))))...--------- ( -31.80, z-score = -2.74, R) >dp4.chrXR_group8 2417312 99 + 9212921 CCCAUCCGCUCGCUGU--CUCAGCGCACUUCGCUUUGCUCACUUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAAAGCAGACA------------ ..........(((((.--..)))))......((((((((.....)))((..((((.(((((((((.....)))))))))))))..))....))))).....------------ ( -28.60, z-score = -1.84, R) >droPer1.super_36 511560 99 - 818889 CCCAUCCGCUAGCUGU--CUCAGCGCACUUCGCUUUGCUCACUUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAAAGCAGACA------------ ....((.(((.((((.--..))))(((...((((..(....)..))))...((((.(((((((((.....))))))))))))).)))......))).))..------------ ( -28.70, z-score = -1.77, R) >consensus __________CCCUGUCGCUCUGCGUAUUUCGCUUUGCUCACUUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAGCACAGCCCC_____CCCAA_ ............((((......(((.....)))...(((.....)))((..((((.(((((((((.....)))))))))))))..)).......))))............... (-21.67 = -21.76 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:54 2011