| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,627,078 – 13,627,192 |
| Length | 114 |
| Max. P | 0.991550 |

| Location | 13,627,078 – 13,627,192 |
|---|---|
| Length | 114 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.90 |
| Shannon entropy | 0.42625 |
| G+C content | 0.45311 |
| Mean single sequence MFE | -32.21 |
| Consensus MFE | -19.52 |
| Energy contribution | -20.05 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
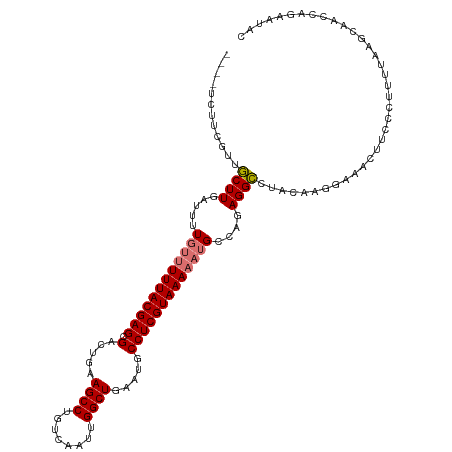
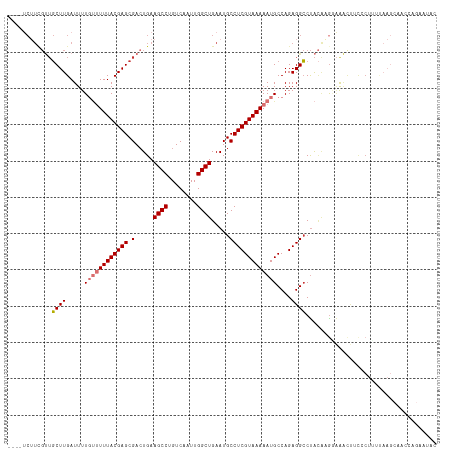
>dm3.chr3L 13627078 114 - 24543557 ----UCUUCGUUGCUUGAUUUUGUUUUUACGAGCGACUGAAGCCUGUCAAUUGGCUGAAUGCCUCGUAAAAAUGCCAGAGGCCUACAAGGAAACGUCCCUUUUAAGCAACCAGAAUAC ----(((..(((((((((....(((((((((((.(.....((((........)))).....))))))))))))(((...)))....((((.......))))))))))))).))).... ( -34.40, z-score = -3.14, R) >droSim1.chr3L 13010956 114 - 22553184 ----UCUUCGUUGCUUGAUUUUGUUUUUACGAGCGACUGAAGCCUGUCAAUUGGCUGAAUGCCUCGUAAAAAUGCCAGAGGCCUACAAAAAAACUUCCCUUUUAAGCAACCAGAAUAC ----(((..(((((((((....(((((((((((.(.....((((........)))).....))))))))))))(((...)))...................))))))))).))).... ( -33.00, z-score = -3.80, R) >droSec1.super_0 5789080 114 - 21120651 ----UCUUCGUUGCUUGAUUUUGUUUUUACGAGCGACUGAAGCCUGUCAAUUGGCUGAAUGCCUCGUAAAAAUGCCAGAGGCCUACAAGGAAACUUCCCUUUUAAGCAACCAGAAUAC ----(((..(((((((((....(((((((((((.(.....((((........)))).....))))))))))))....((((.....(((....))).))))))))))))).))).... ( -35.90, z-score = -3.85, R) >droYak2.chr3L 13721210 114 - 24197627 ----UCUUCGUUGCUUGAUUUUGUUUUUACGAGCGACUGAAGCCUGUCAAUUGGCUGAAUGCCUCGUAAAAAUGCCAGAGGCCUACAUGCAAACUUUCCUUUUAAGCAACCAGAAUGA ----(((..(((((((((....(((((((((((.(.....((((........)))).....))))))))))))(((...)))...................))))))))).))).... ( -33.00, z-score = -2.87, R) >droEre2.scaffold_4784 13629717 114 - 25762168 ----UCUUCGUUGCUUGAUUUUGUUUUUACGAGCGACUGAAGCCUGUCAAUUGGCUGAAUGCCUCGUAAAAAUGCCAGAGGCCUACAUGCAAACUUUCCUUUUAAGCAGCCAGAAUAC ----(((..(((((((((....(((((((((((.(.....((((........)))).....))))))))))))(((...)))...................))))))))).))).... ( -33.00, z-score = -2.68, R) >droAna3.scaffold_13337 1479555 102 + 23293914 ----UCUUCGUUGCUUGAUUUUGCUUUUACGAGCGACUGAAGCCUGUCAAUUGGCUGAAUGCCUCGUAAAAAUGCCAGAGGCCUACGAGGAGAGUUCGCUUCAAAG------------ ----.....((((((((............))))))))((((((..(((....))).((((.(((((((.....(((...))).)))))))...))))))))))...------------ ( -33.40, z-score = -2.22, R) >droWil1.scaffold_180698 10833045 88 + 11422946 ------------UCUUGAUUUU---UUUACGAGCGACUGAAGCCUGUCAAUUGGCUGAAUGCCUCGUAAAAAUGCCAGAGGGCCGUCCGGGGCUCCCUCACAC--------------- ------------...((...((---((((((((.(.....((((........)))).....)))))))))))...))(((((((......)))..))))....--------------- ( -28.10, z-score = -1.57, R) >droGri2.scaffold_15110 17572033 94 - 24565398 UCUUGCGUUGCUGCUUGAUUUU-UCUUUACGAGCGAGUGAAGCCUGUCAAUUGGCUGAAUGCCUCGUAAAAAUGCCAGAGGCCUAGACGGCGGUC----------------------- (((.(((((.......((....-))((((((((.(.((..((((........))))..)).)))))))))))))).))).(((.....)))....----------------------- ( -26.90, z-score = -0.28, R) >consensus ____UCUUCGUUGCUUGAUUUUGUUUUUACGAGCGACUGAAGCCUGUCAAUUGGCUGAAUGCCUCGUAAAAAUGCCAGAGGCCUACAAGGAAACUUCCCUUUUAAGCAACCAGAAUAC ............((((.....((((((((((((.(.....((((........)))).....)))))))))))))....)))).................................... (-19.52 = -20.05 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:53 2011