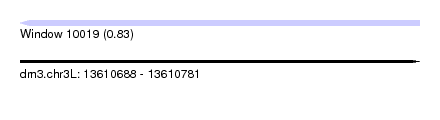
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,610,688 – 13,610,781 |
| Length | 93 |
| Max. P | 0.834707 |

| Location | 13,610,688 – 13,610,781 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 76.77 |
| Shannon entropy | 0.49007 |
| G+C content | 0.40408 |
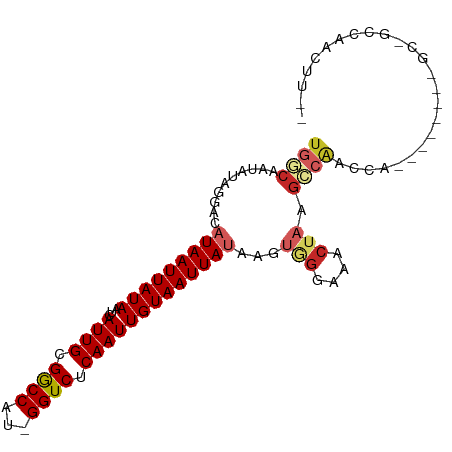
| Mean single sequence MFE | -20.65 |
| Consensus MFE | -13.61 |
| Energy contribution | -14.57 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
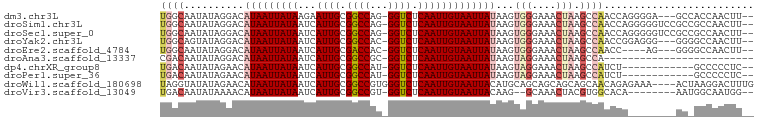
>dm3.chr3L 13610688 93 - 24543557 UGGCAAUAUAGGACAUAAUUAUAAGAAUUGCGGCCAG-GGUCUCAAUUGUAAUUAUAAGUGGGAAACUAAGCCAACCAGGGGA---GCCACCAACUU-- ((((..........(((((((((...((((.(((...-.))).)))))))))))))...(((....))).))))....((...---.))........-- ( -23.00, z-score = -1.01, R) >droSim1.chr3L 12994336 96 - 22553184 UGGCAAUAUAGGACAUAAUUAUAAUCAUUGCGGCCAG-GGUCUCAAUUGUAAUUAUAAGUGGGAAACUAAGCCAACCAGGGGGUCCGCCGCCAACUU-- ((((..........(((((((((((...((.(((...-.))).)))))))))))))...(((....))).))))....((.((....)).)).....-- ( -27.30, z-score = -1.21, R) >droSec1.super_0 5772398 96 - 21120651 UGGCAAUAUAGGACAUAAUUAUAAUCAUUGCGGCCAG-GGUCUCAAUUGUAAUUAUAAGUGGGAAACUAAGCCAACCAGGGGGUCCGCCGCCAACUU-- ((((..........(((((((((((...((.(((...-.))).)))))))))))))...(((....))).))))....((.((....)).)).....-- ( -27.30, z-score = -1.21, R) >droYak2.chr3L 13703692 93 - 24197627 UGGCAGUAUAGGACAUAAUUAUAAUCAUUGCGGCCAC-GGUCUCAAUUGUAAUUAUAAGUGGGAAACUAAGCCAACCGGAGGG---GGGGCCAACUU-- ((((..........(((((((((((...((.(((...-.))).)))))))))))))...(((....)))..((..((...)).---.))))))....-- ( -23.90, z-score = -0.88, R) >droEre2.scaffold_4784 13611504 89 - 25762168 UGGCAAUAUAGGACAUAAUUAUAAUCAUUGCGACCAC-GGUCUCAAUUGUAAUUAUAAGUGGGAAACUAAGCCAACC----AG---GGGGCCAACUU-- ((((..(...((..(((((((((((...((.(((...-.))).)))))))))))))...(((....)))......))----..---)..))))....-- ( -22.40, z-score = -1.80, R) >droAna3.scaffold_13337 1463285 73 + 23293914 CGACAAUAUAGGACAUAAUUAUAAUCAUUGCGGCCGC-GGUCUCAAUUGUAAUUAUAAGUAGGAAACUAAGCCA------------------------- ..........((.((((((((((((...((.(((...-.))).)))))))))))))...(((....))).))).------------------------- ( -16.50, z-score = -2.08, R) >dp4.chrXR_group8 2390492 84 + 9212921 UGACAAUAUAGAACAUAAUUAUAAUCAUUGCGGCCAU-GGUCUCAAUUGUAAUUAUAAGUAGGAAACUAAGCCAUCU------------GCCCCCUC-- ........((((..(((((((((((...((.(((...-.))).)))))))))))))...(((....))).....)))------------).......-- ( -16.20, z-score = -1.27, R) >droPer1.super_36 484778 84 - 818889 UGACAAUAUAGAACAUAAUUAUAAUCAUUGCGGCCAU-GGUCUCAAUUGUAAUUAUAAGUAGGAAACUAAGCCAUCU------------GCCCCCUC-- ........((((..(((((((((((...((.(((...-.))).)))))))))))))...(((....))).....)))------------).......-- ( -16.20, z-score = -1.27, R) >droWil1.scaffold_180698 610849 95 - 11422946 UAGGUAUAUAGAACAUAAUUAUAAUCAUUGCGGCCGUGGGUCUCAAUUGUAAUUACAUGCAGCAGCAGCAGCAACAGAGAAA----ACUAAGGACUUUG .((((...(((....((((((((((...((.((((...)))).))))))))))))..(((.((....)).))).........----.)))...)))).. ( -16.80, z-score = 0.22, R) >droVir3.scaffold_13049 5411709 86 + 25233164 UGACAAUAUAAAACAUAAUUAUAAUCAUUGCGGCCGU-GGUCUCAAUUGUAAUUACAAG--GCAAACUACGUGGCACA--------AAUGGCAAUGG-- .........................(((((((.((((-(((.....((((....)))).--....)))))).).).(.--------...))))))).-- ( -16.90, z-score = -0.95, R) >consensus UGGCAAUAUAGGACAUAAUUAUAAUCAUUGCGGCCAU_GGUCUCAAUUGUAAUUAUAAGUGGGAAACUAAGCCAACCA________GC_GCCAACUU__ ((((..........(((((((((...((((.((((...)))).)))))))))))))...(((....))).))))......................... (-13.61 = -14.57 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:52 2011