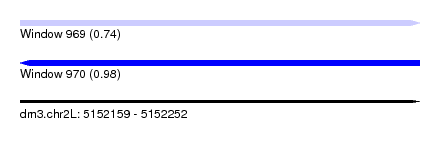
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,152,159 – 5,152,252 |
| Length | 93 |
| Max. P | 0.978478 |

| Location | 5,152,159 – 5,152,252 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 62.24 |
| Shannon entropy | 0.74931 |
| G+C content | 0.50337 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -12.26 |
| Energy contribution | -11.74 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.44 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.737655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
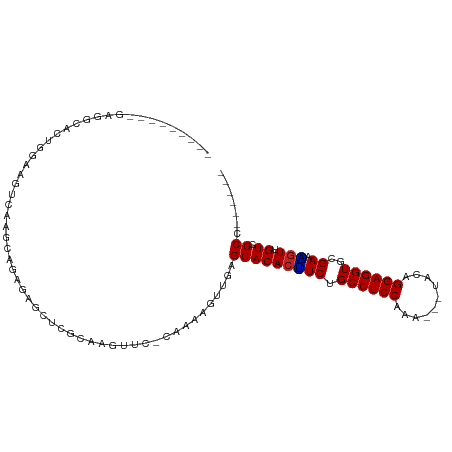
>dm3.chr2L 5152159 93 + 23011544 ---------GAGGCACUGGAAGUCAAGCAGAGUGCUCGCAAGUUC-CAAAAGUUGAGCACACUUCUGCCUCCAAAAGUUACAGGAGGUCCGAAAGUUGUCGCC------ ---------..(((...(((......(((((((((((((......-.....).)))))))...)))))((((..........)))).)))((......)))))------ ( -25.80, z-score = -0.57, R) >droWil1.scaffold_180772 7779461 89 - 8906247 --------UAGUAAACCAAAAGAAAAGUCAGGGUUUUUUUUUUUUGUUUUGUGUAAGCACACAUCUGCCUCCUGU------AGGAGGUGGGAUUGUUGUGGCU------ --------...((((.(((((((((((........))))))))))).))))....(((.((((((..(((((...------.)))))..)).....)))))))------ ( -26.80, z-score = -2.27, R) >dp4.chr4_group3 8945900 91 - 11692001 -----------------AGAAGAGCUAGAAGGAGGAGGAGGAGAGGCUCUUGUUGAGCACACGUCUGCCUCCGGG-CCAAAAGGAGGUGCGAACGUAGUCGCCAGCAGU -----------------......(((....((.(((((((..(.(((((.....)))).).)..)).)))))...-)).......(((((.......).)))))))... ( -29.50, z-score = -0.40, R) >droAna3.scaffold_12916 6172051 86 - 16180835 -------------GAAGGAAAUUCAAGCAGUGAGCUCGGGAGCUC-CAACAGUUGAGCACACUUCUGCCUCCGGG---CCAAGGAGGUGGGACGUUUGUCGCC------ -------------...((.....(((((.(((.((((((..(...-...)..)))))).)))(((..(((((...---....)))))..))).)))))...))------ ( -29.70, z-score = -0.46, R) >droEre2.scaffold_4929 5233323 92 + 26641161 ---------GAGGCACUGGAAGUCAAGCAGAGAGCUAGCAAGUUG-CAAAAGUUGAGCACACUUCUGCCUCCAAA-GUUACAGGAGGUCCGAAAGUUGUCGCC------ ---------..((((((((((((..(((.....))).((.....)-).............))))))((((((...-......)))))).....)).))))...------ ( -21.70, z-score = 0.91, R) >droYak2.chr2L 11952373 92 - 22324452 ---------GAGGCACUGGAAGUCAAGCAGAGAGCUCGCAGGUUC-CAAAAGUUGAGCACACUUCUGCCUCCAAA-GUCACAGGAGGUCCGAAAGUUGUCGCC------ ---------..((((((((((((...((.((....))))..((((-........))))..))))))((((((...-......)))))).....)).))))...------ ( -22.60, z-score = 0.98, R) >droSec1.super_5 3238634 92 + 5866729 ---------GAGGCACUGGAAGUCAAGCAGAGUGCUCGCAAGUUC-CAAAAGUUGAGCACACUUCUGCCUCCAAA-GUUACAGGAGGUCCGAAAGUUGUCGCC------ ---------..(((...(((......(((((((((((((......-.....).)))))))...)))))((((...-......)))).)))((......)))))------ ( -25.90, z-score = -0.51, R) >droSim1.chr2L 4959769 92 + 22036055 ---------GAGGCACUGGAAGUCAAGCAGAGUGCUCGCAAGUUC-CAAAAGUUGAGCACACUUCUGCCUCCAAA-GUUACAGGAGGUCCGAAAGUUGUCGCC------ ---------..(((...(((......(((((((((((((......-.....).)))))))...)))))((((...-......)))).)))((......)))))------ ( -25.90, z-score = -0.51, R) >droGri2.scaffold_15252 7519048 79 - 17193109 ----------------------AUAAAUAAACGAAGCGCGAUUAAACAAUUGUUAAGCACACGUCUGCCUCCCCA---GAGAGGAGGUGCGACCGUUGUCGCUA----- ----------------------........(((..((((((((....))))))...))...)))..((((((...---....))))))(((((....)))))..----- ( -19.70, z-score = -1.06, R) >droMoj3.scaffold_6500 6761604 99 - 32352404 AACGAAUUAAAAAUAAAACAAAGUUGAGGAAUAAGAAGCAGUUAA-CAAUUGUUAAGCACACGUCUGCCUCCCCA---GUCAGGAGGUGCGAUUGUUGUCGCU------ ....................................(((...(((-(((((((...((....))..((((((...---....))))))))))))))))..)))------ ( -19.00, z-score = -0.11, R) >droVir3.scaffold_12963 13761230 79 - 20206255 -------------------AACACAAACAAGC-ACAAGAAGUUAA-CAAUUGUUAAGCACACGUCUGCCUCCGCA---GUCAGGAGGUGCGACUGUUGUCGCU------ -------------------..........(((-((((.(..((((-(....)))))......((((((((((...---....))))))).)))).)))).)))------ ( -21.00, z-score = -0.85, R) >consensus _________GAGGCACUGGAAGUCAAGCAGAGAGCUCGCAAGUUC_CAAAAGUUGAGCACACUUCUGCCUCCAAA___UACAGGAGGUGCGAAAGUUGUCGCC______ ........................................................(((((((((.((((((..........))))))..)).))).)).))....... (-12.26 = -11.74 + -0.53)

| Location | 5,152,159 – 5,152,252 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 62.24 |
| Shannon entropy | 0.74931 |
| G+C content | 0.50337 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -13.23 |
| Energy contribution | -13.08 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
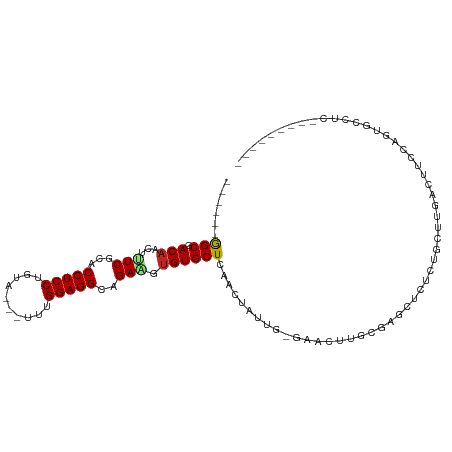
>dm3.chr2L 5152159 93 - 23011544 ------GGCGACAACUUUCGGACCUCCUGUAACUUUUGGAGGCAGAAGUGUGCUCAACUUUUG-GAACUUGCGAGCACUCUGCUUGACUUCCAGUGCCUC--------- ------(((((((..((((.(.(((((..........)))))).))))..)).)).....(((-(((....(((((.....)))))..)))))).)))..--------- ( -28.80, z-score = -1.28, R) >droWil1.scaffold_180772 7779461 89 + 8906247 ------AGCCACAACAAUCCCACCUCCU------ACAGGAGGCAGAUGUGUGCUUACACAAAACAAAAAAAAAAAACCCUGACUUUUCUUUUGGUUUACUA-------- ------(((.(((...(((...(((((.------...)))))..))).))))))...................(((((..((......))..)))))....-------- ( -16.90, z-score = -1.28, R) >dp4.chr4_group3 8945900 91 + 11692001 ACUGCUGGCGACUACGUUCGCACCUCCUUUUGG-CCCGGAGGCAGACGUGUGCUCAACAAGAGCCUCUCCUCCUCCUCCUUCUAGCUCUUCU----------------- ...((((((((......))))..........((-...(((((.((..(.(.((((.....))))).)..))))))).))...))))......----------------- ( -24.30, z-score = -0.52, R) >droAna3.scaffold_12916 6172051 86 + 16180835 ------GGCGACAAACGUCCCACCUCCUUGG---CCCGGAGGCAGAAGUGUGCUCAACUGUUG-GAGCUCCCGAGCUCACUGCUUGAAUUUCCUUC------------- ------((.(((....))))).(((((....---...)))))..(((((..((((........-))))...(((((.....))))).)))))....------------- ( -25.60, z-score = -0.27, R) >droEre2.scaffold_4929 5233323 92 - 26641161 ------GGCGACAACUUUCGGACCUCCUGUAAC-UUUGGAGGCAGAAGUGUGCUCAACUUUUG-CAACUUGCUAGCUCUCUGCUUGACUUCCAGUGCCUC--------- ------(((((......))(((.((((......-...))))((((((((.......)))))))-)......(.(((.....))).)...)))...)))..--------- ( -24.00, z-score = -0.11, R) >droYak2.chr2L 11952373 92 + 22324452 ------GGCGACAACUUUCGGACCUCCUGUGAC-UUUGGAGGCAGAAGUGUGCUCAACUUUUG-GAACCUGCGAGCUCUCUGCUUGACUUCCAGUGCCUC--------- ------(((((((..((((.(.(((((......-...)))))).))))..)).)).....(((-(((....(((((.....)))))..)))))).)))..--------- ( -29.10, z-score = -0.89, R) >droSec1.super_5 3238634 92 - 5866729 ------GGCGACAACUUUCGGACCUCCUGUAAC-UUUGGAGGCAGAAGUGUGCUCAACUUUUG-GAACUUGCGAGCACUCUGCUUGACUUCCAGUGCCUC--------- ------(((((((..((((.(.(((((......-...)))))).))))..)).)).....(((-(((....(((((.....)))))..)))))).)))..--------- ( -28.90, z-score = -1.28, R) >droSim1.chr2L 4959769 92 - 22036055 ------GGCGACAACUUUCGGACCUCCUGUAAC-UUUGGAGGCAGAAGUGUGCUCAACUUUUG-GAACUUGCGAGCACUCUGCUUGACUUCCAGUGCCUC--------- ------(((((((..((((.(.(((((......-...)))))).))))..)).)).....(((-(((....(((((.....)))))..)))))).)))..--------- ( -28.90, z-score = -1.28, R) >droGri2.scaffold_15252 7519048 79 + 17193109 -----UAGCGACAACGGUCGCACCUCCUCUC---UGGGGAGGCAGACGUGUGCUUAACAAUUGUUUAAUCGCGCUUCGUUUAUUUAU---------------------- -----..(((((....))))).((((((...---..)))))).(((((.((((..(((....))).....))))..)))))......---------------------- ( -27.00, z-score = -3.07, R) >droMoj3.scaffold_6500 6761604 99 + 32352404 ------AGCGACAACAAUCGCACCUCCUGAC---UGGGGAGGCAGACGUGUGCUUAACAAUUG-UUAACUGCUUCUUAUUCCUCAACUUUGUUUUAUUUUUAAUUCGUU ------.((((......))))..........---(((((((..(((...((..(((((....)-))))..)).)))..)))))))........................ ( -18.60, z-score = -0.22, R) >droVir3.scaffold_12963 13761230 79 + 20206255 ------AGCGACAACAGUCGCACCUCCUGAC---UGCGGAGGCAGACGUGUGCUUAACAAUUG-UUAACUUCUUGU-GCUUGUUUGUGUU------------------- ------.(((((....))))).(((((....---...)))))((((((.(..((((((....)-))))......).-.).))))))....------------------- ( -24.50, z-score = -1.24, R) >consensus ______GGCGACAACUUUCGCACCUCCUGUA___UUUGGAGGCAGAAGUGUGCUCAACUAUUG_GAACUUGCGAGCUCUCUGCUUGACUUCCAGUGCCUC_________ ......(((.(((...(((...(((((..........)))))..))).))))))....................................................... (-13.23 = -13.08 + -0.15)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:11 2011