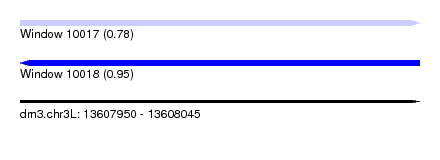
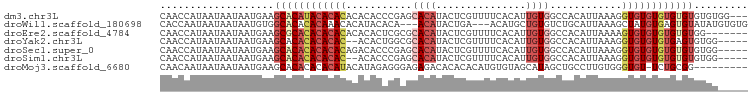
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,607,950 – 13,608,045 |
| Length | 95 |
| Max. P | 0.953251 |

| Location | 13,607,950 – 13,608,045 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 75.52 |
| Shannon entropy | 0.48651 |
| G+C content | 0.42912 |
| Mean single sequence MFE | -30.91 |
| Consensus MFE | -13.41 |
| Energy contribution | -15.57 |
| Covariance contribution | 2.16 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782150 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
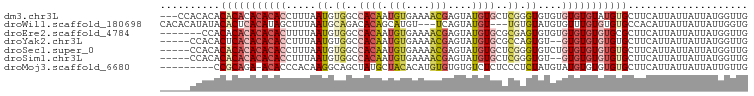
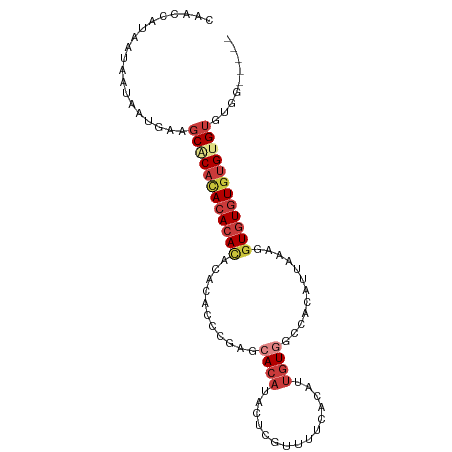
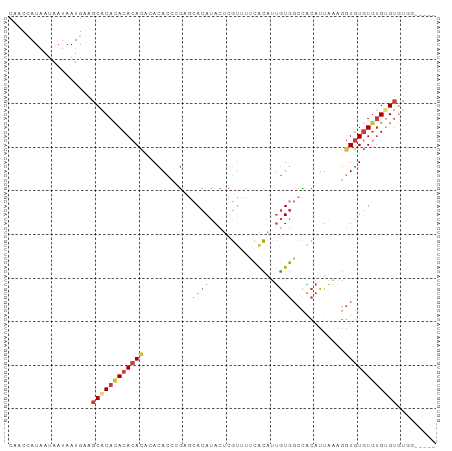
>dm3.chr3L 13607950 95 + 24543557 ---CCACACACACACACACACCUUUAAUGUGGCCACAAUGUGAAAACGAGUAUGUGCUCGGGUGUGUGUGUGUAUGUGCUUCAUUAUUAUUAUGGUUG ---.((((.(((((((((((((((((...((....))...))))..(((((....)))))))))))))))))).)))).................... ( -36.40, z-score = -2.88, R) >droWil1.scaffold_180698 10807309 92 - 11422946 CACACAUAUACACUCACAUAGCUUUAAUGCAGACACAGCAUGU---UCAGUAUGU---UGUGUAUGUGUUUGUGUGUGCCACAUUAUUAUUAUUGGUG ((((((((.((((.......((......))..((((((((((.---....)))))---)))))..)))).))))))))...(((((.......))))) ( -29.00, z-score = -1.57, R) >droEre2.scaffold_4784 13608786 91 + 25762168 -------CCACACACACACACUUUUAAUGUGGCCACAAUGUGAAAACGAGUAUGUGCGCGAGUGUGUGUGUGUGUGCGCUUCAUUAUUAUUAUGGUUG -------.(((((((((((((((........((((((.(((....)))....)))).)))))))))))))))))........................ ( -30.40, z-score = -1.37, R) >droYak2.chr3L 13700923 91 + 24197627 -----CCACACUCACACACACCUUUAAUGUGGCCACAAUGUGAAAACGAGUAUGUGCGCCAGUGU--GUGUGUGUGUGCUUCAUUAUUAUUAUGGUUG -----.(((((.(((((((((........((((((((.(((....)))....)))).))))))))--))))).))))).................... ( -32.50, z-score = -2.56, R) >droSec1.super_0 5769649 93 + 21120651 -----CCACACACACACACACCUUUAAUGUGGCCACAAUGUGAAAACGAGUAUGUGCUCGGGUGUCUGUGUGUGUGUGCUUCAUUAUUAUUAUGGUUG -----.((((((((((((((((((((...((....))...))))..(((((....))))))))))..))))))))))).................... ( -32.70, z-score = -2.22, R) >droSim1.chr3L 12991467 91 + 22553184 -----CCACACACACACACACCUUUAAUGUGGCCACAAUGUGAAAACGAGUAUGUGCUCGGGUGU--GUGUGUGUGUGCUUCAUUAUUAUUAUGGUUG -----.((((((((((((((((((((...((....))...))))..(((((....))))))))))--))))))))))).................... ( -38.10, z-score = -3.90, R) >droMoj3.scaffold_6680 17243675 88 - 24764193 ---------CCGCAGA-ACACCCACAAGGCAGCUAUGCUACACAUGUGUGUGUCUCUCCCUCUAUGUAUGUGUGUGUGCUUCAUUAUUAUUAUUGUUG ---------..(.((.-((((.((((.((((....))))(((((....)))))...............)))).)))).)).)................ ( -17.30, z-score = 0.62, R) >consensus _____CCACACACACACACACCUUUAAUGUGGCCACAAUGUGAAAACGAGUAUGUGCUCGGGUGUGUGUGUGUGUGUGCUUCAUUAUUAUUAUGGUUG ..........(((((((((((.....((.((..((((.(((....)))....))))..)).))....))))))))))).................... (-13.41 = -15.57 + 2.16)
| Location | 13,607,950 – 13,608,045 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 75.52 |
| Shannon entropy | 0.48651 |
| G+C content | 0.42912 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -13.46 |
| Energy contribution | -14.89 |
| Covariance contribution | 1.43 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953251 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
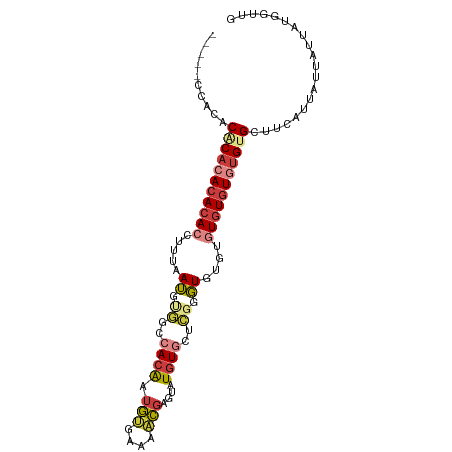
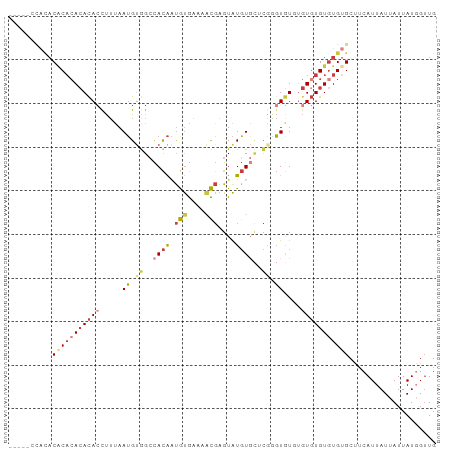
>dm3.chr3L 13607950 95 - 24543557 CAACCAUAAUAAUAAUGAAGCACAUACACACACACACCCGAGCACAUACUCGUUUUCACAUUGUGGCCACAUUAAAGGUGUGUGUGUGUGUGUGG--- ....................((((((((((((((((((.(((......)))((..((((...))))..))......)))))))))))))))))).--- ( -37.70, z-score = -4.23, R) >droWil1.scaffold_180698 10807309 92 + 11422946 CACCAAUAAUAAUAAUGUGGCACACACAAACACAUACACA---ACAUACUGA---ACAUGCUGUGUCUGCAUUAAAGCUAUGUGAGUGUAUAUGUGUG ...............((((.....)))).((((((((((.---(((((((..---..((((.......))))...)).)))))..)))))..))))). ( -22.00, z-score = 0.02, R) >droEre2.scaffold_4784 13608786 91 - 25762168 CAACCAUAAUAAUAAUGAAGCGCACACACACACACACUCGCGCACAUACUCGUUUUCACAUUGUGGCCACAUUAAAAGUGUGUGUGUGUGG------- ........................((((((((((((((.((.((((...............)))))).........)))))))))))))).------- ( -26.96, z-score = -1.97, R) >droYak2.chr3L 13700923 91 - 24197627 CAACCAUAAUAAUAAUGAAGCACACACACAC--ACACUGGCGCACAUACUCGUUUUCACAUUGUGGCCACAUUAAAGGUGUGUGUGAGUGUGG----- ....................(((((.(((((--((((((((.((((...............)))))))........)))))))))).))))).----- ( -30.56, z-score = -2.63, R) >droSec1.super_0 5769649 93 - 21120651 CAACCAUAAUAAUAAUGAAGCACACACACACAGACACCCGAGCACAUACUCGUUUUCACAUUGUGGCCACAUUAAAGGUGUGUGUGUGUGUGG----- ......................((((((((((.(((((.(((......)))((..((((...))))..))......))))).)))))))))).----- ( -31.70, z-score = -2.82, R) >droSim1.chr3L 12991467 91 - 22553184 CAACCAUAAUAAUAAUGAAGCACACACACAC--ACACCCGAGCACAUACUCGUUUUCACAUUGUGGCCACAUUAAAGGUGUGUGUGUGUGUGG----- ....................(((((((((((--(((((.(((......)))((..((((...))))..))......)))))))))))))))).----- ( -35.30, z-score = -4.15, R) >droMoj3.scaffold_6680 17243675 88 + 24764193 CAACAAUAAUAAUAAUGAAGCACACACACAUACAUAGAGGGAGAGACACACACAUGUGUAGCAUAGCUGCCUUGUGGGUGU-UCUGCGG--------- ...................(((...((((...((((.(((.((.(..((((....))))..)....)).))))))).))))-..)))..--------- ( -19.60, z-score = -0.36, R) >consensus CAACCAUAAUAAUAAUGAAGCACACACACACACACACCCGAGCACAUACUCGUUUUCACAUUGUGGCCACAUUAAAGGUGUGUGUGUGUGUGG_____ ...................((((((((((((...........((((...............))))............))))))))))))......... (-13.46 = -14.89 + 1.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:51 2011