
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,578,082 – 13,578,180 |
| Length | 98 |
| Max. P | 0.820018 |

| Location | 13,578,082 – 13,578,180 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 75.48 |
| Shannon entropy | 0.50365 |
| G+C content | 0.38250 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -14.04 |
| Energy contribution | -13.83 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
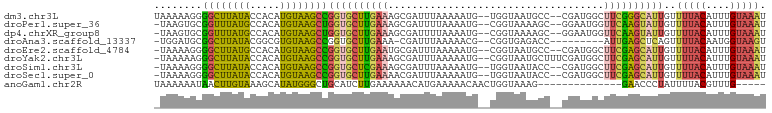
>dm3.chr3L 13578082 98 + 24543557 UAAAAAGGGGCUUAUACCACAUGUAAGCCGGUGCUUGAAAGCGAUUUAAAAAUG--UGGUAAUGCC--CGAUGGCUUCGGGCAUUGUUUUACAUUUGUAAAU ........((((((((.....))))))))..((((....))))(((((.(((((--((((((((((--(((.....))))))))))..)))))))).))))) ( -34.40, z-score = -3.75, R) >droPer1.super_36 449034 97 + 818889 -UAAGUGCGGUUUAUGCCACAUGUAAGCUGGUGCUUGAAAGCGAUUUUAAAAUG--CGGUAAAAGC--GGAAUGGUUCAAGUAUUGUUUUACAUUUGUAAAU -....((((((....)))..(((((((..(((((((((..((.((((.....((--(.......))--))))).)))))))))))..)))))))..)))... ( -22.30, z-score = -0.68, R) >dp4.chrXR_group8 2354465 97 - 9212921 -UAAGUGCGGUUUAUGCCACAUGUAAGCUGGUGCUUGAAAGCGAUUUUAAAAUG--CGGUAAAAGC--GGAAUGGUUCAAGUAUUGUUUUACAUUUGUAAAU -....((((((....)))..(((((((..(((((((((..((.((((.....((--(.......))--))))).)))))))))))..)))))))..)))... ( -22.30, z-score = -0.68, R) >droAna3.scaffold_13337 1426212 89 - 23293914 -UGGAUGCGGCUUAUACGGCGUGUAAGCCGGUGCUUGAAA-CGAUUUAAAAACG--CGGUGAGACC---------AUUGAGCUCAGUUUUACAAUGGUAAGU -....((((((((((((...))))))))).((..(((((.-...)))))..)))--)).....(((---------((((............))))))).... ( -22.50, z-score = -0.17, R) >droEre2.scaffold_4784 13578182 97 + 25762168 -UAAAAGGGGCUUAUGCCACAUGUAAGCCGGUGCUUGAAUGCGAUUUAAAAAUG--CGGUAAUGCC--CGAUGGCUUCGAGCAUUGUUUUACAUUUGUAAAU -.......(((....)))..(((((((.(((((((((((.((.((.........--(((......)--)))).))))))))))))).)))))))........ ( -28.50, z-score = -1.52, R) >droYak2.chr3L 13670243 99 + 24197627 -UAAAAAGGGCUUAUACCACAUGUAAGCCGGUGCUUGAAAGCGAUUUAAAAAUG--CGGUAAUGCUUUCGAUGGCUUCGAGCAUUGUUUUACAUUUGUAAAU -.......((((((((.....))))))))((((((((((.((.(((.(((...(--((....)))))).))).))))))))))))..(((((....))))). ( -29.00, z-score = -2.71, R) >droSim1.chr3L 12956569 97 + 22553184 -UAAAAGGGGCUUAUACCACAUGUAAGCCGGUGCUCGAAAGCGAUUUAAAAAUG--UGGUAAUACC--CGAUGGCUUCGAGCAUUGUUUUACAUUUGUAAAU -.......((((((((.....))))))))((((((((((.((.(((.......(--.((.....))--)))).))))))))))))..(((((....))))). ( -28.61, z-score = -2.66, R) >droSec1.super_0 5740086 97 + 21120651 -UAAAAGGGGCUUAUACCACAUGUAAGCCGGUGCUUGAAAACGAUUUAAAAAUG--UGGUAAUACC--CGAUGGCUUCGAGCAUUGUUUUACAUUUGUAAAU -.......((((((((.....))))))))((((((((((..((...........--(((......)--)).))..))))))))))..(((((....))))). ( -23.10, z-score = -1.21, R) >anoGam1.chr2R 12235123 83 + 62725911 UAAAAAAUAACUUGUAAAGCAUAUGGGCUGCAUCUUGAAAAAACAUGAAAAACAACUGGUAAAG--------------GAACCCUAUUUUACGUUUG----- .......(((..(((..(((......))))))..)))...((((.((.....))....((((((--------------........)))))))))).----- ( -6.40, z-score = 1.55, R) >consensus _UAAAAGGGGCUUAUACCACAUGUAAGCCGGUGCUUGAAAGCGAUUUAAAAAUG__CGGUAAUACC__CGAUGGCUUCGAGCAUUGUUUUACAUUUGUAAAU ........((((((((.....))))))))((((((((((....................................))))))))))..(((((....))))). (-14.04 = -13.83 + -0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:45 2011