
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,568,265 – 13,568,395 |
| Length | 130 |
| Max. P | 0.839614 |

| Location | 13,568,265 – 13,568,395 |
|---|---|
| Length | 130 |
| Sequences | 4 |
| Columns | 139 |
| Reading direction | reverse |
| Mean pairwise identity | 48.27 |
| Shannon entropy | 0.80717 |
| G+C content | 0.57959 |
| Mean single sequence MFE | -44.88 |
| Consensus MFE | -13.22 |
| Energy contribution | -14.21 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.839614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
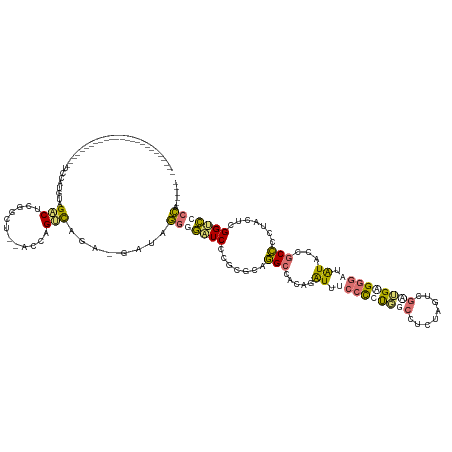
>dm3.chr3L 13568265 130 - 24543557 UCCAUCAACUGAAAGAUGGCCUACGGUUACCUAGGCGAAGC---ACCAGCCUUA-GAAAGGGGUUCCCACGUAGGCCACGGAUUUCCCCCUGGCUUUACGGAUGGAGGUUUGUACCGCCUCGACUCGGACACCA----- ((((((.....((((.(((((((((.....((((((.....---....))).))-)...(((...))).)))))))))(((........))).))))...))))))(((..((.(((........)))))))).----- ( -43.10, z-score = -0.48, R) >droPer1.super_11490 445 127 + 901 ------UCCAAAGAGGUUUCCUACCUCCAUGUCG-CUUGCUUUACCAGGUCAGGCGAUAGGGGAUCCCGCGCUGGCCACAGGUUUCCCUUGGCUUCUGGUCGAUGGGGGAUAUACCGCCCCUUCUCGGUCCCCA----- ------......(((((.....)))))..(((((-((((((......)).)))))))))(((((((((.((.((((((.(((((......))))).)))))).)).)))))..((((........)))))))).----- ( -52.40, z-score = -1.86, R) >dp4.Unknown_group_286 28769 109 - 32412 -------------------------UCCAGGGAGACACCACUUAACCAGUCAGGCGAUAGGGGAUCCCGCGCUGGCCACAGGUUUCCCUUGGCUUCUGGUCGGUGAGGGAUAUACCGCCCCUUUUCGGUCCCCA----- -------------------------....(((.(((.........((.....))(((.((((((((((.(((((((((.(((((......))))).))))))))).)))))......)))))..))))))))).----- ( -46.90, z-score = -1.86, R) >droWil1.scaffold_181113 16559 107 + 65715 ---------------------------AUUAUUGACUCGGCC--GCGCGUUCUA-GAGGCAAUUUCCCUCGCAAGACAAGAAUUUCCUCUUGCCGCAA--CGCUGCGAGUUAUGCCGCUCCAACUCGGCGGCGACACCA ---------------------------.....(((((((((.--(((.((...(-((((.(((((...((....))...))))).))))).)))))..--.))..)))))))(((((((.......)))))))...... ( -37.10, z-score = -2.20, R) >consensus _________________________UCCAUGUAGACUCGGCU__ACCAGUCAGA_GAUAGGGGAUCCCGCGCAGGCCACAGAUUUCCCCUGGCCUCUAGUCGAUGAGGGAUAUACCGCCCCUACUCGGUCCCCA_____ .................................(((............)))........(((((((.......(((.....((.(((((((((........))))))))).))...))).......)))))))...... (-13.22 = -14.21 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:43 2011