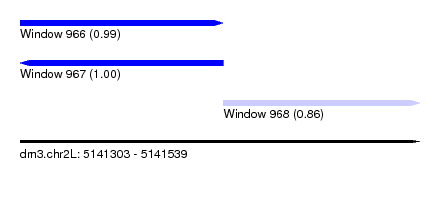
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,141,303 – 5,141,539 |
| Length | 236 |
| Max. P | 0.998859 |

| Location | 5,141,303 – 5,141,423 |
|---|---|
| Length | 120 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.42 |
| Shannon entropy | 0.51250 |
| G+C content | 0.50314 |
| Mean single sequence MFE | -34.54 |
| Consensus MFE | -18.75 |
| Energy contribution | -18.67 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
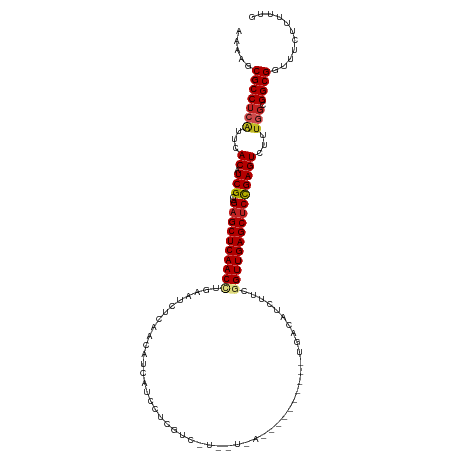
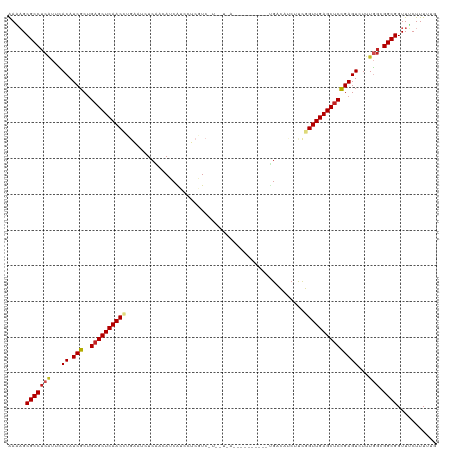
>dm3.chr2L 5141303 120 + 23011544 CACAAAGAAACCGCCUCCAAAGACUCGGAGCUCAACCGGAGGUGUCAGCACAGAUGAUGAUAAAGACGAGGCUGAUGUUGAGAUUCAGGUUGAGCUCAGCGAUGUGAAUGAGGCGCUUUU ......((((.((((((.....(((((((((((((((.(((.(.(((((((((....((.......))...))).)))))).)))).))))))))))..))).))....)))))).)))) ( -42.40, z-score = -2.94, R) >droGri2.scaffold_15252 7506228 90 - 17193109 UAAAAAGCAACCGCCGCCAAAUACUCAGAGCUCAACCGACGAUGACA------------------A------CGAUCUCGAC------GUUGAGCUCAGCGAUGUGAACGAGGCGCUAUU ............((.(((.........((((((((((((.(((....------------------.------..))))))..------))))))))).....((....)).))))).... ( -25.40, z-score = -2.18, R) >droMoj3.scaffold_6500 6748213 90 - 32352404 UAUAAAGAAACCGCCGCCAAAGACUCAGAGCUCAACCGAGGAUGACA------------------A------CGAUCUCGAC------GUUGAGCUCAGCGAUGUGAAUGAGGCGCUGUC ......((...((((..((...((((.((((((((((((((.((...------------------.------)).)))))..------)))))))))...)).))...)).))))...)) ( -26.80, z-score = -1.75, R) >droVir3.scaffold_12963 13747442 96 - 20206255 UAAAAAGCAACCGCCGCCAAAUACUCAGAGCUCAACCGAAGACGACA------------------AUGAGAACGAUCUCGAC------GUUGAGCUCAGCGAUGUGAAUGAGGCGCUGUU .....((((..((((..((..(((((.(((((((((((....))...------------------.(((((....)))))..------)))))))))...)).)))..)).)))).)))) ( -28.00, z-score = -2.25, R) >droPer1.super_1 6036696 102 - 10282868 GAAAAAGCAACCGCCGCAAAAGACUCGGAGCUCAACCGACGAUAUC------------------GAUGAAGAUGAUGAUGAGAUCCAAGUUGAGCUCAGCGAUGUCCAUGAGGCGCGUUU ........(((((((.((...(((((((((((((((...((.((((------------------......)))).)).((.....)).)))))))))..))).)))..)).)))).))). ( -30.90, z-score = -2.12, R) >dp4.chr4_group3 8930328 102 - 11692001 GAAAAAGCAACCGCCGCAAAAGACUCGGAGCUCAACCAACGAUAUC------------------GAUGAAGAUGAUGAUGAGAUCCAAGUUGAGCUCAGCGAUGUCCAUGAGGCGCGUUU ........(((((((.((...(((((((((((((((...((.((((------------------......)))).)).((.....)).)))))))))..))).)))..)).)))).))). ( -30.90, z-score = -2.36, R) >droAna3.scaffold_12916 6160104 114 - 16180835 UGUAAAGAAACCGCCGCCAAAGACUCGGGGCUCAACCGAAGAUGUCGGGCG------UCAGGAGGAUGAAAACGAUGCCGAGAUUCAGGUUGAGCUCGGCGAUGUGAAGGAGGCGCUUUU ......((((.((((.((....(((((((((((((((...(((.((((.((------((..............)))))))).)))..))))))))))..))).))...)).)))).)))) ( -41.84, z-score = -1.90, R) >droEre2.scaffold_4929 5222379 120 + 26641161 CUAAAAGAAACCGCCUCCAAAGACUCGGAGCUCAACCGAAGGUGUCAACAGGAAUGAUGAUAAGGACGAGGAUGAUGUUGAGAUUCAGGUUGAGCUCAGCGAUGUGCAUGAGGCGCUUUC ......((((.((((((.....(((((((((((((((...(((.((((((.........................)))))).)))..))))))))))..))).))....)))))).)))) ( -40.41, z-score = -2.77, R) >droYak2.chr2L 11941429 120 - 22324452 CAUAAAGAAACCGCCUCCAAAGACUCGGAGCUCAACCAAAGGUGUCAACAGAGAUGAUGAUGAAGACGAGGAUGAUGUUGAGAUUCAGGUUGAGCUCAGCGAUGUAAAUGAGGCGCUUUU ......((((.((((((.....(((((((((((((((...(((.((((((.........................)))))).)))..))))))))))..))).))....)))))).)))) ( -38.91, z-score = -3.35, R) >droSec1.super_5 3228028 120 + 5866729 CACAAAGAAACCGCCUGCAAAGACUCGGAGCUCAACCGGAGGUGUCAACAGAGAUGAUGAUGAGGACGAGGAUGAUGUUGAGAUUCAGGUUGAGCUCAGCGAUGUGAAUGAGGCGCUUUU ......((((.(((((.((...(((((((((((((((.(((.(.((((((.........................)))))).)))).))))))))))..))).))...))))))).)))) ( -37.61, z-score = -1.92, R) >droSim1.chr2L 4949128 120 + 22036055 CCCAAAGAAACCGCCUCCAAAGACUCGGAGCUCAACCAGAGGUGUCAACCGAGAUGAUGAUGAGGACGAGGAUGAUGUUGAGAUUCAGGUUGAGCUCAGCGAUGUGAAUGAGGCGCUUUU ......((((.((((((.....(((((((((((((((...(((....)))..........((((..(((........)))...))))))))))))))..))).))....)))))).)))) ( -36.80, z-score = -1.54, R) >consensus CAAAAAGAAACCGCCGCCAAAGACUCGGAGCUCAACCGAAGAUGUCA__________U_A__A_GACGAGGAUGAUGUUGAGAUUCAGGUUGAGCUCAGCGAUGUGAAUGAGGCGCUUUU ...........((((.......((((.(((((((((....................................................)))))))))...)).))......))))..... (-18.75 = -18.67 + -0.08)

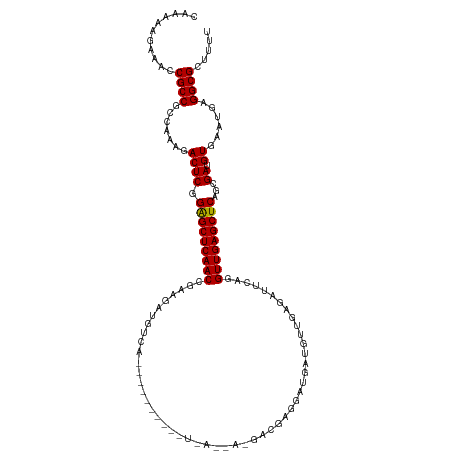
| Location | 5,141,303 – 5,141,423 |
|---|---|
| Length | 120 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.42 |
| Shannon entropy | 0.51250 |
| G+C content | 0.50314 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.998859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5141303 120 - 23011544 AAAAGCGCCUCAUUCACAUCGCUGAGCUCAACCUGAAUCUCAACAUCAGCCUCGUCUUUAUCAUCAUCUGUGCUGACACCUCCGGUUGAGCUCCGAGUCUUUGGAGGCGGUUUCUUUGUG .(((.((((((.......(((..((((((((((((.....))...((((((..................).))))).......))))))))))))).......)))))).)))....... ( -38.41, z-score = -3.07, R) >droGri2.scaffold_15252 7506228 90 + 17193109 AAUAGCGCCUCGUUCACAUCGCUGAGCUCAAC------GUCGAGAUCG------U------------------UGUCAUCGUCGGUUGAGCUCUGAGUAUUUGGCGGCGGUUGCUUUUUA ...(((((((((((.....(...(((((((((------(.((((((..------.------------------.))).))).).)))))))))...).....))))).)).))))..... ( -29.40, z-score = -1.15, R) >droMoj3.scaffold_6500 6748213 90 + 32352404 GACAGCGCCUCAUUCACAUCGCUGAGCUCAAC------GUCGAGAUCG------U------------------UGUCAUCCUCGGUUGAGCUCUGAGUCUUUGGCGGCGGUUUCUUUAUA ((.(.(((((((...((.(((..(((((((((------..((((....------.------------------.......))))))))))))))))))...))).)))).).))...... ( -28.70, z-score = -1.27, R) >droVir3.scaffold_12963 13747442 96 + 20206255 AACAGCGCCUCAUUCACAUCGCUGAGCUCAAC------GUCGAGAUCGUUCUCAU------------------UGUCGUCUUCGGUUGAGCUCUGAGUAUUUGGCGGCGGUUGCUUUUUA ..((((((((((...((.(((..(((((((((------...((((....))))..------------------...((....))))))))))))))))...))).))).))))....... ( -30.40, z-score = -1.58, R) >droPer1.super_1 6036696 102 + 10282868 AAACGCGCCUCAUGGACAUCGCUGAGCUCAACUUGGAUCUCAUCAUCAUCUUCAUC------------------GAUAUCGUCGGUUGAGCUCCGAGUCUUUUGCGGCGGUUGCUUUUUC .(((.((((.((.((((.(((..(((((((((...(((......)))........(------------------(((...))))))))))))))))))))..)).)))))))........ ( -34.10, z-score = -2.73, R) >dp4.chr4_group3 8930328 102 + 11692001 AAACGCGCCUCAUGGACAUCGCUGAGCUCAACUUGGAUCUCAUCAUCAUCUUCAUC------------------GAUAUCGUUGGUUGAGCUCCGAGUCUUUUGCGGCGGUUGCUUUUUC .(((.((((.((.((((.(((..((((((((((..(((.((...............------------------)).)))...)))))))))))))))))..)).)))))))........ ( -34.76, z-score = -3.02, R) >droAna3.scaffold_12916 6160104 114 + 16180835 AAAAGCGCCUCCUUCACAUCGCCGAGCUCAACCUGAAUCUCGGCAUCGUUUUCAUCCUCCUGA------CGCCCGACAUCUUCGGUUGAGCCCCGAGUCUUUGGCGGCGGUUUCUUUACA .(((.((((.((......(((..(.((((((((.(((..((((...((((...........))------)).))))....)))))))))))).)))......)).)))).)))....... ( -34.70, z-score = -1.97, R) >droEre2.scaffold_4929 5222379 120 - 26641161 GAAAGCGCCUCAUGCACAUCGCUGAGCUCAACCUGAAUCUCAACAUCAUCCUCGUCCUUAUCAUCAUUCCUGUUGACACCUUCGGUUGAGCUCCGAGUCUUUGGAGGCGGUUUCUUUUAG ((((.((((((.......(((..((((((((((.(((..((((((.........................))))))....)))))))))))))))).......)))))).))))...... ( -41.55, z-score = -4.71, R) >droYak2.chr2L 11941429 120 + 22324452 AAAAGCGCCUCAUUUACAUCGCUGAGCUCAACCUGAAUCUCAACAUCAUCCUCGUCUUCAUCAUCAUCUCUGUUGACACCUUUGGUUGAGCUCCGAGUCUUUGGAGGCGGUUUCUUUAUG .(((.((((((.......(((..((((((((((......((((((.........................)))))).......))))))))))))).......)))))).)))....... ( -37.07, z-score = -4.19, R) >droSec1.super_5 3228028 120 - 5866729 AAAAGCGCCUCAUUCACAUCGCUGAGCUCAACCUGAAUCUCAACAUCAUCCUCGUCCUCAUCAUCAUCUCUGUUGACACCUCCGGUUGAGCUCCGAGUCUUUGCAGGCGGUUUCUUUGUG .(((.(((((((...((.(((..((((((((((.((...((((((.........................))))))....)).)))))))))))))))...)).))))).)))....... ( -35.71, z-score = -3.77, R) >droSim1.chr2L 4949128 120 - 22036055 AAAAGCGCCUCAUUCACAUCGCUGAGCUCAACCUGAAUCUCAACAUCAUCCUCGUCCUCAUCAUCAUCUCGGUUGACACCUCUGGUUGAGCUCCGAGUCUUUGGAGGCGGUUUCUUUGGG .(((.((((((.......(((..((((((((((.((...(((((...........................)))))....)).))))))))))))).......)))))).)))....... ( -37.17, z-score = -2.51, R) >consensus AAAAGCGCCUCAUUCACAUCGCUGAGCUCAACCUGAAUCUCAACAUCAUCCUCGUC_U__U_A__________UGACAUCUUCGGUUGAGCUCCGAGUCUUUGGCGGCGGUUUCUUUUUG .....(((((((...((.(((..((((((((((..................................................)))))))))))))))...))).))))........... (-22.37 = -22.72 + 0.36)
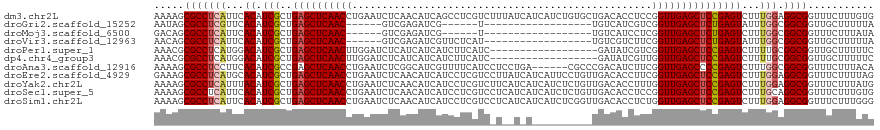
| Location | 5,141,423 – 5,141,539 |
|---|---|
| Length | 116 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.89 |
| Shannon entropy | 0.47533 |
| G+C content | 0.49400 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -21.30 |
| Energy contribution | -20.00 |
| Covariance contribution | -1.30 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.856485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
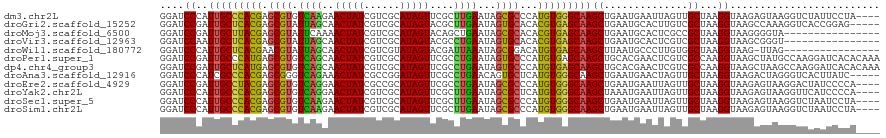
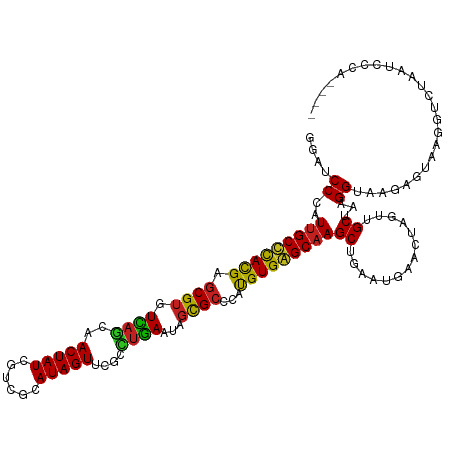
>dm3.chr2L 5141423 116 + 23011544 GGAUCCCAUUGCCCACGAGCGUGUCAAGAACUAUCGUCGCAUAGUUCGCUUGAAUAGCGCCCAUGUGGGCAAGCUGAAUGAAUUAGUUGCUAAGGUAAGAGUAAGGUCUAUUCCUA---- (((((...(((((((((.((((.(((.(((((((......)))))))...)))...))))...)))))))))(((((.....)))))((((........)))).))))).......---- ( -35.40, z-score = -1.80, R) >droGri2.scaffold_15252 7506318 115 - 17193109 GGAUCCGAUUGCUCACGAGCGUAUUAGCAACUAUCGUCGCAUAGUACGCUUGAAUAGUGCACACGUGAGCAAGCUGAAUGCACUUGUCGCUAAGGUAAGCCAAAGGUCACCGGAG----- ...((((.((((((((((((((((((((.((....)).)).)))))))))).....((....))))))))))((.....))((((...(((......)))...))))...)))).----- ( -38.60, z-score = -1.98, R) >droMoj3.scaffold_6500 6748303 104 - 32352404 GGAUCCGAUUGCUUACGAGCGUAUUCAAAACUAUCGUCGCAUAGUACAGCUGAAUAGCGCACACGUGAGCAAGCUGAAUGCACUCGCCGCUAAGGUAAGGGGUA---------------- ..((((..(((((((((.(((((((((..(((((......))))).....))))).))))...)))))))))((.....)).((..((.....))..)))))).---------------- ( -32.10, z-score = -0.95, R) >droVir3.scaffold_12963 13747538 104 - 20206255 GGAUCCAAUUGCUCACGAGCGUAUUAGCAACUAUCGUCGCAUAGUACGCCUGAAUAGUGCACACGUGAGCAAGCUGAAUGCACUCGUCGCUAAGGUAAGCGGGU---------------- (.((.((.(((((((((.((((((((((.((....)).)).)))))))).......(....).)))))))))..)).)).)((((((.((....))..))))))---------------- ( -33.10, z-score = -0.90, R) >droWil1.scaffold_180772 7764801 103 - 8906247 GGAUCCCAUUGCUCACGAACGUAUUAGCAACUAUCGUCGUAUAGUACGAUUAAAUAGCGGACAUGUGAGCAAGCUUAAUGCCCUUGUGGCUAAGGUAAG-UUAG---------------- .........((((((((...((....((....((((((.....).)))))......))..)).))))))))((((((..(((.....))).....))))-))..---------------- ( -23.90, z-score = -0.11, R) >droPer1.super_1 6036798 120 - 10282868 GGAUCCGAUUGCCCAUGAGCGUGUCAGCAACUAUCGUCGCAUAGUUCGCCUGAAUAGUGCCCAUGUGAGCAAGCUGCACGAACUCGUCGCCAAGGUAAGCUAUGCCAAGGAUCACACAAA .(((((..(((.(.(((((((((.((((..........((((.((((....)))).))))...((....)).))))))))..))))).).)))((((.....))))..)))))....... ( -35.20, z-score = -0.80, R) >dp4.chr4_group3 8930430 120 - 11692001 GGAUCCGAUUGCUCAUGAGCGUGUCAGCAACUAUCGUCGCAUAGUUCGCCUGAAUAGUGCCCAUGUGAGCAAGCUGCACGAACUCGUCGCCAAGGUAAGCUAAGCCAAGGAUCACACAAA .(((((..(((((((((.((((.(((((((((((......)))))).).))))...)))).))..)))))))...(((((....))).))...(((.......)))..)))))....... ( -33.70, z-score = -0.33, R) >droAna3.scaffold_12916 6160218 115 - 16180835 GGAUCCCAUCGCCCACGAGCGGGUCAGAAACUAUCGCCGGAUAGUUCGCCUGAACAGUGCUCAUGUGGGCAAGCUGAAUGAACUAGUUGCUAAGGUAAGACUAGGGUCACUUAUC----- .(((((....(((((((((((.(((((.(((((((....)))))))...))).))..)))))..)))))).............(((((((....))..)))))))))).......----- ( -37.70, z-score = -1.28, R) >droEre2.scaffold_4929 5222499 116 + 26641161 GGAUCCGAUUGCCUACGAGCGUGUCAGGAACUAUCGCCGCAUAGUUCGCCUGAAUAGCGCCCAUGUGGGCAAGCUGAAUGAAUUAGUUGCUAAGGUAAGAGUAAGGACUAUCCCCA---- ((((((..((((((...(((((.(((((.(((((......)))))...)))))...))((((....)))).((((((.....))))))))).))))))......)))...)))...---- ( -36.70, z-score = -1.46, R) >droYak2.chr2L 11941549 116 - 22324452 GGAUCCCAUUGCCCACGAGCGUGUCAGGAACUAUCGUCGCAUAGUUCGCUUGAAUAGCGCUCAUGUGGGCAAGCUAAAUGAAUUAGUUGCUAAGGUAAGAGUAAGGUUCAUCCCCA---- ((......((((((((((((((.(((((((((((......))))))).).)))...))))))..)))))))).....(((((((..(((((........))))))))))))..)).---- ( -40.80, z-score = -3.31, R) >droSec1.super_5 3228148 116 + 5866729 GGAUCCCAUUGCCCACGAGCGUGUCAAGAACUAUCGUCGCAUAGUUCGCUUGAAUAGCGCCCAUGUGGGCAAGCUGAAUGAAUUAGUUGCUAAGGUAAGAGUAAGGUCUAAUCCUA---- ((((.((.(((((((((.((((.(((.(((((((......)))))))...)))...))))...)))))))))(((((.....)))))((((........)))).))....))))..---- ( -36.00, z-score = -2.07, R) >droSim1.chr2L 4949248 116 + 22036055 GGAUCCCAUUGCCCACGAGCGUGUCAAGAACUAUCGUCGCAUAGUUCGCUUGAAUAGCGCCCAUGUGGGCAAGCUGAAUGAAUUAGUUGCUAAGGUAAGAGUAAGGUCUAAUCCUA---- ((((.((.(((((((((.((((.(((.(((((((......)))))))...)))...))))...)))))))))(((((.....)))))((((........)))).))....))))..---- ( -36.00, z-score = -2.07, R) >consensus GGAUCCCAUUGCCCACGAGCGUGUCAGCAACUAUCGUCGCAUAGUUCGCCUGAAUAGCGCCCAUGUGAGCAAGCUGAAUGAACUAGUUGCUAAGGUAAGAGUAAGGUCUAAUCCCA____ ....((..(((((((((.((((.((((..(((((......)))))....))))...))))...)))))))))((..............))...))......................... (-21.30 = -20.00 + -1.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:18:10 2011