
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,548,191 – 13,548,317 |
| Length | 126 |
| Max. P | 0.514269 |

| Location | 13,548,191 – 13,548,317 |
|---|---|
| Length | 126 |
| Sequences | 12 |
| Columns | 135 |
| Reading direction | forward |
| Mean pairwise identity | 86.43 |
| Shannon entropy | 0.29515 |
| G+C content | 0.37168 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -21.99 |
| Energy contribution | -22.15 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.514269 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 13548191 126 + 24543557 -----UGGAUGUGGAGCUUUAAUGC-CUUGUCU--GCAUUUGUUUUCAACUGUCCGUGCGUGCGCCAUUUUAAUUAACAUGCAAAUCCAGUUGAAAAUGCCAAUUUAUGCUUGACAUAAUAUACCGAGAAAAAA- -----...((((.((((...(((((-.......--))))).(((((((((((....((((((...............))))))....)))))))))))..........)))).)))).................- ( -30.96, z-score = -1.77, R) >droSim1.chr3L_random 629905 126 + 1049610 -----UGGAUGUGGAGCUUUAAUGC-CUUGUCU--GCAUUUGUUUUCAACUGUCCGUGCGUGCGCCAUUUUAAUUAACAUGCAAAUCCAGUUGAAAAUGCCAAUUUAUGCUUGACAUAAUAUACCGAGAAAAAA- -----...((((.((((...(((((-.......--))))).(((((((((((....((((((...............))))))....)))))))))))..........)))).)))).................- ( -30.96, z-score = -1.77, R) >droSec1.super_0 5715806 126 + 21120651 -----UGGAUGUGGAGUUUUAAUGC-UUUGUCU--GCAUUUGUUUUCAACUGUCCGUGCGUGCGCCAUUUUAAUUAACAUGCAAAUCCAGUUGAAAAUGCCAAUUUAUGCUUGACAUAAUAUACCGAGAAAAAA- -----...((((.((((...(((((-.......--))))).(((((((((((....((((((...............))))))....)))))))))))..........)))).)))).................- ( -29.86, z-score = -1.41, R) >droYak2.chr3L 13644042 127 + 24197627 -----UGGAUGUGGAGCUUUAAUGC-CUUGUCU--GCAUUUGUUUUCAACUGUCCGUGCGUGCGCCAUUUUAAUUAACAUGCAAAUCCAGUUGAAAAUGCCAAUUUAUGCUUGACAUAAUAUACCGAAAAAAAAA -----...((((.((((...(((((-.......--))))).(((((((((((....((((((...............))))))....)))))))))))..........)))).)))).................. ( -30.96, z-score = -2.05, R) >droEre2.scaffold_4784 13553489 126 + 25762168 -----UGGAUGUGGAGCUUUAAUGC-CUUGUCU--GCAUUUGUUUUCAACUGUCCGUGCGUGCGCCAUUUUAAUUAACAUGCAAAUCCAGUUGAAAAUGCCAAUUUAUGCUUGACAUAAUAUACCGAGAAAAAA- -----...((((.((((...(((((-.......--))))).(((((((((((....((((((...............))))))....)))))))))))..........)))).)))).................- ( -30.96, z-score = -1.77, R) >droAna3.scaffold_13337 1401051 129 - 23293914 -UCAGAGUGGGCCG-GCUUUAAUGC-CUUGUCU--GCAUUUGUUUUCAACUGUCCGUGCGUGCGCCAUUUUAAUUAACAUGCAAAUCCAGUUGAAAAUGCCAAUUUAUGCUUGACAUAAUAUACCGAGAAAAAA- -......(((...(-((......))-).((((.--((((..(((((((((((....((((((...............))))))....)))))))))))........))))..)))).......)))........- ( -31.96, z-score = -1.33, R) >dp4.chrXR_group8 2325610 129 - 9212921 CGGAGAGGGAGACGAGCUUUAAUGC-CGUGUCU--GAAUUUGCUUUCCACUGUCCGUGCGUGCGCCAUUUUAAUUAACAUGCAAAUCCAGUUGAAAAUGCCAAUUUAUGCUUGACAUAAUAUACAGAAAACA--- .(..((((((.(((.((......))-))).)))--(((((.((((((.((((....((((((...............))))))....)))).))))..)).)))))...)))..).................--- ( -28.56, z-score = -0.17, R) >droPer1.super_36 420425 129 + 818889 CGGAGAGAGAGACGAGCUUUAAUGC-CGUGUCU--GAAUUUGCUUUCCACUGUCCGUGCGUGCGCCAUUUUAAUUAACAUGCAAAUCCAGUUGAAAAUGCCAAUUUAUGCUUGACAUAAUAUACAGAAAACA--- .(..((((((.(((.((......))-))).)))--(((((.((((((.((((....((((((...............))))))....)))).))))..)).)))))...)))..).................--- ( -28.36, z-score = -0.71, R) >droWil1.scaffold_180698 10733710 125 - 11422946 -----UGUAGAGGGGGCUUUAAUGC-CUUGUCUGC-CAUUUGUUUUCAACUGUCCGUGCGUGCGCCAUUUUAAUUAACAUGCAAAUCCAGUUGAAAAUGCCAAUUUAUGCUUGACAUAAUAUACAAAAAGCA--- -----.(((((.(((((......))-))).)))))-.....(((((((((((....((((((...............))))))....))))))))))).........(((((...............)))))--- ( -36.82, z-score = -2.68, R) >droVir3.scaffold_13049 5342518 129 - 25233164 -----GUUAUUGGGUGCUUUAAUGCCCUUGUCUCUGCAUUUGUUUUCAGCUGUCCGUGCGUGCAGCAUUUUAAUUAACAUGCAAAUCCAGUUGAAA-UGCCAAUUUAUGCUUGACAUAUAAUAUACAAAAAAAUA -----(((((.((((........)))).((((...(((((((.(((((((((....((((((...............))))))....)))))))))-...)))...))))..)))).)))))............. ( -30.56, z-score = -1.45, R) >droMoj3.scaffold_6680 17170730 119 - 24764193 ------------UAUGCUUUAAUGCUCUUGUCUCUGCAUUUGUUUUCAGCUGUCCGUGCGUGCAGCAUUUUAAUUAACAUGCAAAUCCAGUUGAAA-UGCCAAUUUAUGCUUGACAUAUAAUA-ACAAAAAAA-- ------------...((......))...((((...(((((((.(((((((((....((((((...............))))))....)))))))))-...)))...))))..)))).......-.........-- ( -26.26, z-score = -1.40, R) >droGri2.scaffold_15110 17480848 131 + 24565398 ---CAGUUAUUGGGUGCUUUAAUGCUCUUGUCUCGGCAUUUGUUUUCAACUGUCCGUGCGUGCAGCAUUUUAAUUAACAUGCAAAUCCAGUUGAAA-UGCCAAUUUAUGCUUGACAUAUAAUAUACAAAAUAAAA ---..(((((((((.((......)).))((((..((((((((.(((((((((....((((((...............))))))....)))))))))-...)))...))))).))))..))))).))......... ( -29.46, z-score = -1.02, R) >consensus _____UGGAUGUGGAGCUUUAAUGC_CUUGUCU__GCAUUUGUUUUCAACUGUCCGUGCGUGCGCCAUUUUAAUUAACAUGCAAAUCCAGUUGAAAAUGCCAAUUUAUGCUUGACAUAAUAUACAGAAAAAAAA_ ............((.((......)).))((((...(((((((.(((((((((....((((((...............))))))....)))))))))....)))...))))..))))................... (-21.99 = -22.15 + 0.16)
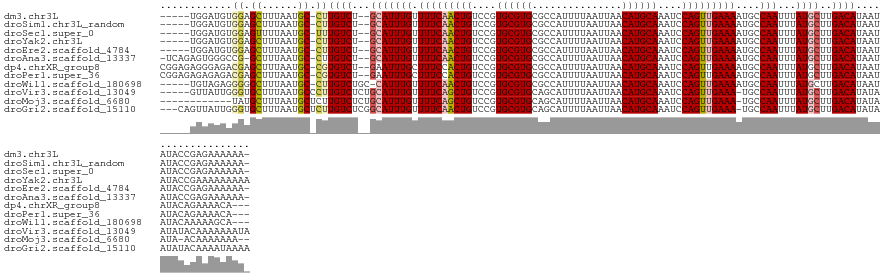
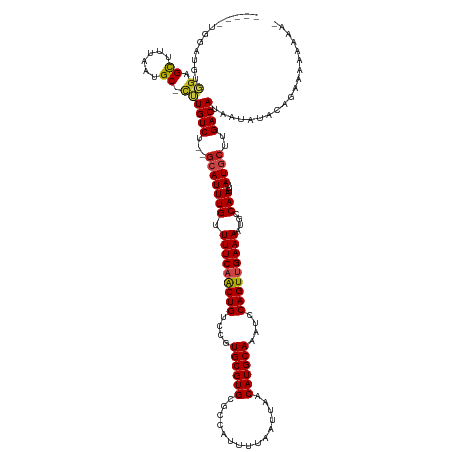
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:41 2011