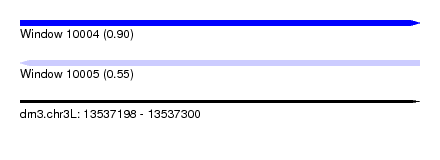
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,537,198 – 13,537,300 |
| Length | 102 |
| Max. P | 0.900988 |

| Location | 13,537,198 – 13,537,300 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 85.08 |
| Shannon entropy | 0.28780 |
| G+C content | 0.62884 |
| Mean single sequence MFE | -50.55 |
| Consensus MFE | -30.72 |
| Energy contribution | -30.75 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
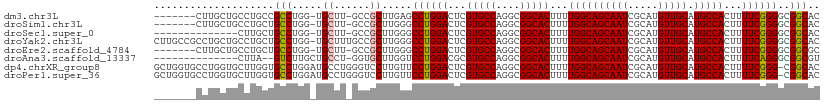
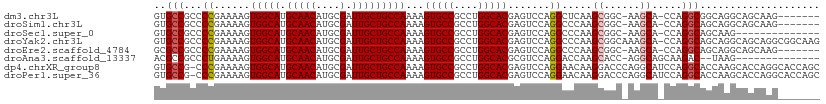
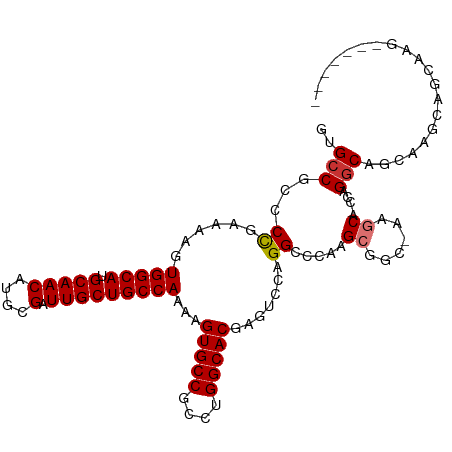
>dm3.chr3L 13537198 102 + 24543557 -------CUUGCUGCCUGCCGCCUGG-UGCUU-GCCGCUUGAGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCGGGGCGGCAC -------..(((((((((((((((((-..(..-(((((....))..)).)..)..))))))))).....((((((((((.....))))).))))).......)))))))). ( -54.60, z-score = -3.81, R) >droSim1.chr3L 3059111 102 - 22553184 -------CUUGCUGCCUGCUGCCUGG-UGCUU-GCCGCUUGGGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCGGGGCGGCAC -------..(((((((((((((((((-..(..-(((((....))..)).)..)..))))))))).....((((((((((.....))))).))))).......)))))))). ( -52.50, z-score = -3.02, R) >droSec1.super_0 5704895 95 + 21120651 --------------CUUGCUGCCUGG-UGCUU-GCCGCUUGGGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCGGGGCGGCAC --------------..((((((((.(-....(-(((((((((((........)).))))))))))....((((((((((.....))))).))))).....).)))))))). ( -45.10, z-score = -2.06, R) >droYak2.chr3L 13632830 110 + 24197627 CUUGCCGCCUGCUGCCUGCUGCCUGG-UGCUUUGCCGCUUGGGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCGGGGCGGCAC ..((((((((((((((((((((((((-..(....((((....))..))....)..)))))))))).....))))))....(((((...))))).........)))))))). ( -58.80, z-score = -3.56, R) >droEre2.scaffold_4784 13542309 102 + 25762168 -------CUUGCUGCCUGCUGCCUGG-UGCUU-GCCGCUUGGGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCGGGGCGGCGC -------...((((((((((((((((-..(..-(((((....))..)).)..)..))))))))).....((((((((((.....))))).))))).......))))))).. ( -51.90, z-score = -2.38, R) >droAna3.scaffold_13337 1388883 94 - 23293914 --------------CUUA--GUCUUGCUGCCU-GGUGCUUGGUCCUGGACGCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCAGGGCGGCGU --------------....--((((((((((((-((..(.((........)).)..))))))))).....((((((((((.....))))).)))))......)))))..... ( -40.90, z-score = -1.47, R) >dp4.chrXR_group8 2313032 110 - 9212921 GCUGGUGCCUGGUGCUUGGUGCCUGGAUGCCUGGGUCCUUGUUCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCGGG-CGGCAC ..((.(((((((((((....((((((.(((..((((((........))))))))))))))))))))...((((((((((.....))))).))))).....))))-)).)). ( -50.30, z-score = -1.77, R) >droPer1.super_36 407802 110 + 818889 GCUGGUGCCUGGUGCUUGGUGCCUGGAUGCCUGGGUCCUUGUUCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCGGG-CGGCAC ..((.(((((((((((....((((((.(((..((((((........))))))))))))))))))))...((((((((((.....))))).))))).....))))-)).)). ( -50.30, z-score = -1.77, R) >consensus _______CCUGCUGCCUGCUGCCUGG_UGCUU_GCCGCUUGGGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCGGGGCGGCAC ....................(((.....((......)).....((((((...(((((....)))))...((((((((((.....))))).)))))...))))))..))).. (-30.72 = -30.75 + 0.03)
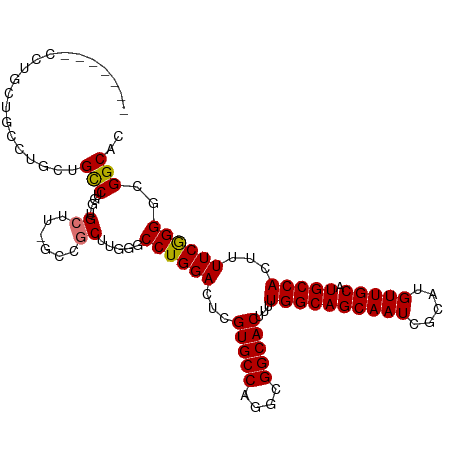
| Location | 13,537,198 – 13,537,300 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 85.08 |
| Shannon entropy | 0.28780 |
| G+C content | 0.62884 |
| Mean single sequence MFE | -43.90 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.36 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.548545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13537198 102 - 24543557 GUGCCGCCCCGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCUCAAGCGGC-AAGCA-CCAGGCGGCAGGCAGCAAG------- (((((((........)))))))((.....))..((((((((.....((((((((((...((((....))))..((...-..)).-)))))))))))))))))).------- ( -53.10, z-score = -3.95, R) >droSim1.chr3L 3059111 102 + 22553184 GUGCCGCCCCGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCCCAAGCGGC-AAGCA-CCAGGCAGCAGGCAGCAAG------- (((((((........)))))))......(((..((((((((....((((.(((((((....).))))))((....)).-..)))-)..))))))))...)))..------- ( -43.80, z-score = -1.76, R) >droSec1.super_0 5704895 95 - 21120651 GUGCCGCCCCGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCCCAAGCGGC-AAGCA-CCAGGCAGCAAG-------------- (((((((........)))))))((.....))..((((((((....((((.(((((((....).))))))((....)).-..)))-)..)))))))).-------------- ( -40.70, z-score = -2.22, R) >droYak2.chr3L 13632830 110 - 24197627 GUGCCGCCCCGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCCCAAGCGGCAAAGCA-CCAGGCAGCAGGCAGCAGGCGGCAAG .(((((((......(((.(((((...))))).)))((((((.....(((((((((((....).))))))....((......)).-...))))...)))))).))))))).. ( -52.30, z-score = -2.50, R) >droEre2.scaffold_4784 13542309 102 - 25762168 GCGCCGCCCCGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCCCAAGCGGC-AAGCA-CCAGGCAGCAGGCAGCAAG------- ..((.(((......(((.(((((...))))).)))((((((....((((.(((((((....).))))))((....)).-..)))-)..)))))).))).))...------- ( -42.40, z-score = -1.16, R) >droAna3.scaffold_13337 1388883 94 + 23293914 ACGCCGCCCUGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGCGUCCAGGACCAAGCACC-AGGCAGCAAGAC--UAAG-------------- ..(((((........))))).(((.....))).((((((((....((((..((((((....).))))).....)))).-.))))))))...--....-------------- ( -35.90, z-score = -2.07, R) >dp4.chrXR_group8 2313032 110 + 9212921 GUGCCG-CCCGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGAACAAGGACCCAGGCAUCCAGGCACCAAGCACCAGGCACCAGC (((((.-.........(((((.(((((....).)))))))))...(((((((((((.....((((........)))))))))).....))))).........))))).... ( -41.50, z-score = -1.57, R) >droPer1.super_36 407802 110 - 818889 GUGCCG-CCCGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGAACAAGGACCCAGGCAUCCAGGCACCAAGCACCAGGCACCAGC (((((.-.........(((((.(((((....).)))))))))...(((((((((((.....((((........)))))))))).....))))).........))))).... ( -41.50, z-score = -1.57, R) >consensus GUGCCGCCCCGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCCCAAGCGGC_AAGCA_CCAGGCAGCAAGCAGCAAG_______ ..(((...((......(((((.(((((....).)))))))))...(((((....))))).......)).....((......)).....))).................... (-25.10 = -25.36 + 0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:40 2011