| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,426,740 – 13,426,839 |
| Length | 99 |
| Max. P | 0.935850 |

| Location | 13,426,740 – 13,426,839 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.04 |
| Shannon entropy | 0.40010 |
| G+C content | 0.56669 |
| Mean single sequence MFE | -33.42 |
| Consensus MFE | -29.11 |
| Energy contribution | -27.91 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
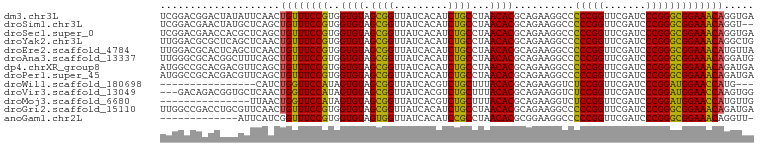
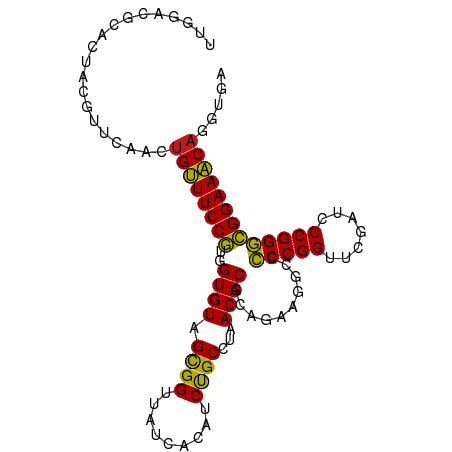
>dm3.chr3L 13426740 99 + 24543557 UCACCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUGAAUAUAGUCCGUCCGA (((.((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))).)))............... ( -34.90, z-score = -1.43, R) >droSim1.chr3L 12829617 97 + 22553184 --ACCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGAGCAUAGUUCGUCCGA --..((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))))..((((...))))...... ( -33.40, z-score = -0.75, R) >droSec1.super_0 5601814 99 + 21120651 UCACCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGAGCGUGGUUCGUCCGA (((.((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))).)))...(((.....))). ( -36.00, z-score = -0.59, R) >droYak2.chr3L 13528768 99 + 24197627 CAGCCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUGAGCUGAGCGCGUCCAA ((((((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))))....))))........... ( -36.30, z-score = -0.91, R) >droEre2.scaffold_4784 13437547 99 + 25762168 UAACAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUGAGCUGAGUGCGUCCAA ((((.(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))))))).((.....))...... ( -33.50, z-score = -0.82, R) >droAna3.scaffold_13337 1846395 99 - 23293914 CAUCCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGAAAGCCGUGCGCCCAA ....((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))))................... ( -31.90, z-score = 0.09, R) >dp4.chrXR_group8 2849465 99 - 9212921 UCAUCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGAACGUCGUGCGGCCAU (((.((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))).)))..(((....)))... ( -36.40, z-score = -1.03, R) >droPer1.super_45 119116 99 + 618639 UCAUCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGAACGUCGUGCGGCCAU (((.((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))).)))..(((....)))... ( -36.40, z-score = -1.03, R) >droWil1.scaffold_180698 7115582 80 - 11422946 ---CAUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGAUG---------------- ---..(((((((((((((.......))))).....(((((.......))))).(((.....))).......))))))))....---------------- ( -28.60, z-score = -3.32, R) >droVir3.scaffold_13049 17111829 96 - 25233164 CCACUUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGUUGAGCACCGUCUGUC--- .((.((((((((((((((.......))))).....(((((.......))))).(((.....))).......))))))))).)).............--- ( -30.40, z-score = -1.50, R) >droMoj3.scaffold_6680 11784331 84 + 24764193 CAACAUGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGUUAA--------------- .(((.(((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))))))..--------------- ( -29.90, z-score = -3.55, R) >droGri2.scaffold_15110 6689306 99 - 24565398 UCAUCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUGAACGCAGGUCGGCCAA (((.((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))).))).....((....)).. ( -36.80, z-score = -1.28, R) >anoGam1.chr2L 19555540 85 - 48795086 -AACCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACCGAUGAAU------------- -...(.((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))).)......------------- ( -29.90, z-score = -0.99, R) >consensus UCACCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUGAACGUAGUGCGUCCAA .....(((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))).................... (-29.11 = -27.91 + -1.20)



| Location | 13,426,740 – 13,426,839 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 82.04 |
| Shannon entropy | 0.40010 |
| G+C content | 0.56669 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -26.69 |
| Energy contribution | -25.56 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.678932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13426740 99 - 24543557 UCGGACGGACUAUAUUCAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGUGA ...............(((.((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).))) ( -33.60, z-score = -0.71, R) >droSim1.chr3L 12829617 97 - 22553184 UCGGACGAACUAUGCUCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGU-- ...................((((((((((((((.......)))))).(((((.......)))))..((((..((......)).))))))))))))..-- ( -32.00, z-score = -0.23, R) >droSec1.super_0 5601814 99 - 21120651 UCGGACGAACCACGCUCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGUGA ..((.....))....(((.((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).))) ( -36.40, z-score = -0.94, R) >droYak2.chr3L 13528768 99 - 24197627 UUGGACGCGCUCAGCUCAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGCUG ...((.((.....))))..((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).... ( -34.70, z-score = -0.62, R) >droEre2.scaffold_4784 13437547 99 - 25762168 UUGGACGCACUCAGCUCAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUGUUA ...((.((.....))))((((((((((((((((.......)))))).(((((.......)))))..((((..((......)).))))))))))).))). ( -32.00, z-score = -0.71, R) >droAna3.scaffold_13337 1846395 99 + 23293914 UUGGGCGCACGGCUUUCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGAUG ..((((.....))))....((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).... ( -33.80, z-score = 0.07, R) >dp4.chrXR_group8 2849465 99 + 9212921 AUGGCCGCACGACGUUCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGAUGA .((..((.....))..)).((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).... ( -33.90, z-score = -0.31, R) >droPer1.super_45 119116 99 - 618639 AUGGCCGCACGACGUUCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGAUGA .((..((.....))..)).((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).... ( -33.90, z-score = -0.31, R) >droWil1.scaffold_180698 7115582 80 + 11422946 ----------------CAUCUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAUG--- ----------------....((((((((..(((((((((.........)))))..))))..........(((((.......)))))))))))))..--- ( -27.70, z-score = -2.41, R) >droVir3.scaffold_13049 17111829 96 + 25233164 ---GACAGACGGUGCUCAACUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAAGUGG ---.............((..((((((((..(((((((((.........)))))..))))..........(((((.......)))))))))))))..)). ( -29.60, z-score = -0.43, R) >droMoj3.scaffold_6680 11784331 84 - 24764193 ---------------UUAACUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCAUGUUG ---------------.((((((((((((..(((((((((.........)))))..))))..........(((((.......))))))))))))).)))) ( -29.60, z-score = -2.81, R) >droGri2.scaffold_15110 6689306 99 + 24565398 UUGGCCGACCUGCGUUCAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGAUGA (((..((.....))..)))((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).... ( -34.70, z-score = -0.88, R) >anoGam1.chr2L 19555540 85 + 48795086 -------------AUUCAUCGGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGUU- -------------....(((.(((((((.(((.(.(((.....))).(((((.......)))))).)))(((((.......)))))))))))).))).- ( -30.60, z-score = -0.85, R) >consensus UUGGACGCACUACGUUCAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGUGA ....................((((((((..((((.((((.........))))...))))..........(((((.......)))))))))))))..... (-26.69 = -25.56 + -1.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:31 2011