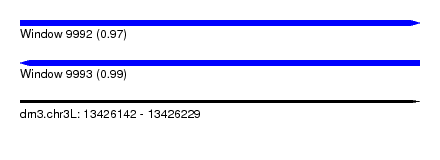
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,426,142 – 13,426,229 |
| Length | 87 |
| Max. P | 0.988153 |

| Location | 13,426,142 – 13,426,229 |
|---|---|
| Length | 87 |
| Sequences | 13 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 85.80 |
| Shannon entropy | 0.31231 |
| G+C content | 0.55732 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -27.11 |
| Energy contribution | -26.57 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13426142 87 + 24543557 CAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGUGAUAAA---------- ((.((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).)).....---------- ( -32.60, z-score = -2.19, R) >droGri2.scaffold_15110 17875141 87 + 24565398 CGACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGAAGCUAAU---------- ...((((((((((((((.......)))))).(((((.......)))))..((((..((......)).))))))))))))........---------- ( -32.10, z-score = -2.15, R) >droMoj3.scaffold_6680 5130360 84 - 24764193 CAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGAUAAC------------- ...((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).....------------- ( -32.10, z-score = -2.81, R) >droVir3.scaffold_13049 17111495 89 - 25233164 CAGCUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGACCCCGGAUGGAACCAUGGCGUAUAAC-------- ..((((((((((..(((((((((.........)))))..))))..........(((((.......))))))))))))..))).......-------- ( -29.50, z-score = -1.44, R) >droWil1.scaffold_180698 7114884 96 - 11422946 CAACUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACCA-GGAUUCUGGUUGAGUCAA ...(((((((((..(((((((((.........)))))..))))..........(((((.......)))))))))))))-)(((((.....))))).. ( -36.30, z-score = -2.96, R) >droPer1.super_45 118515 85 + 618639 CAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACA-UGAUUGG----------- (((((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))-.).))).----------- ( -29.40, z-score = -1.32, R) >dp4.chrXR_group8 2848906 85 - 9212921 CAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACA-UGAUUGG----------- (((((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))-.).))).----------- ( -29.40, z-score = -1.32, R) >droAna3.scaffold_13337 1845583 88 - 23293914 CAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACA--GGAUGAUUAA------- ((.((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))--)..)).....------- ( -32.10, z-score = -1.72, R) >droEre2.scaffold_4784 13437001 84 + 25762168 CAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGUGAU------------- ((.((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).))..------------- ( -32.60, z-score = -2.14, R) >droYak2.chr3L 13528204 85 + 24197627 CAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGUGAAG------------ ((.((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).))...------------ ( -32.60, z-score = -2.21, R) >droSec1.super_0 5600852 87 + 21120651 CAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGUGAUAAA---------- ((.((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).)).....---------- ( -32.60, z-score = -2.19, R) >droSim1.chr3L 12827177 87 + 22553184 CAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGUGAUAAA---------- ((.((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).)).....---------- ( -32.60, z-score = -2.19, R) >anoGam1.chr2L 19552281 84 - 48795086 ---CGGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAAUGGAUGUU---------- ---..(((((((.(((.(.(((.....))).(((((.......)))))).)))(((((.......))))))))))))..........---------- ( -29.90, z-score = -0.34, R) >consensus CAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGGGAUAA___________ ....(((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))................... (-27.11 = -26.57 + -0.54)

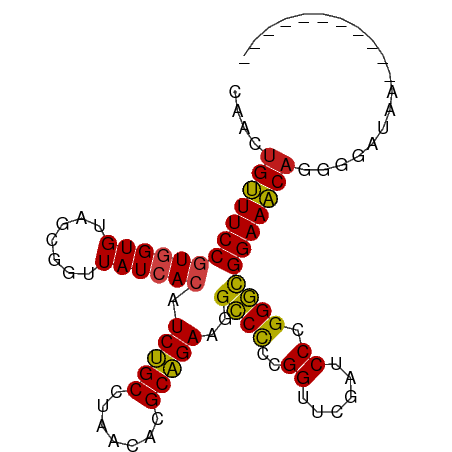
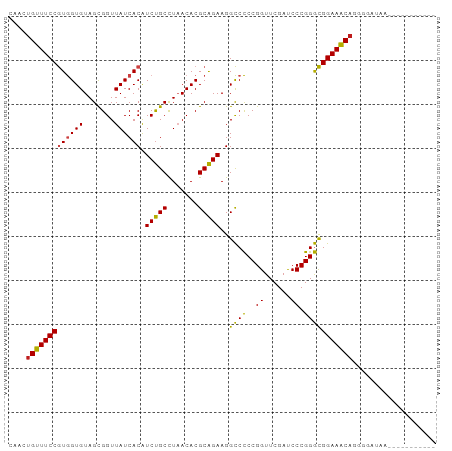
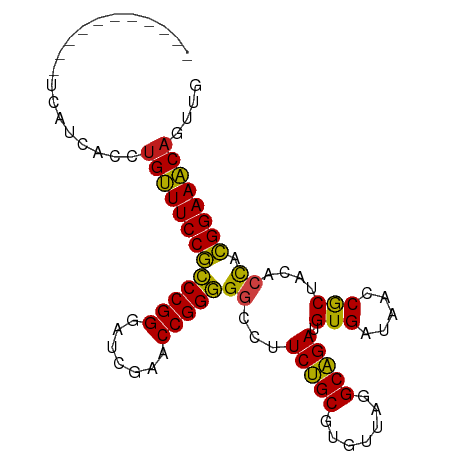
| Location | 13,426,142 – 13,426,229 |
|---|---|
| Length | 87 |
| Sequences | 13 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 85.80 |
| Shannon entropy | 0.31231 |
| G+C content | 0.55732 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -29.48 |
| Energy contribution | -28.58 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13426142 87 - 24543557 ----------UUUAUCACCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUG ----------.....((.((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))).)) ( -32.30, z-score = -2.35, R) >droGri2.scaffold_15110 17875141 87 - 24565398 ----------AUUAGCUUCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUCG ----------........((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))))... ( -31.90, z-score = -1.88, R) >droMoj3.scaffold_6680 5130360 84 + 24764193 -------------GUUAUCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUG -------------.....((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))))... ( -31.90, z-score = -2.22, R) >droVir3.scaffold_13049 17111495 89 + 25233164 --------GUUAUACGCCAUGGUUCCAUCCGGGGUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGCUG --------.......((..(((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))))).. ( -29.60, z-score = -1.95, R) >droWil1.scaffold_180698 7114884 96 + 11422946 UUGACUCAACCAGAAUCC-UGGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGUUG .................(-(((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))))... ( -31.00, z-score = -2.73, R) >droPer1.super_45 118515 85 - 618639 -----------CCAAUCA-UGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUG -----------.((((..-(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))))))) ( -30.00, z-score = -1.57, R) >dp4.chrXR_group8 2848906 85 + 9212921 -----------CCAAUCA-UGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUG -----------.((((..-(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))))))) ( -30.00, z-score = -1.57, R) >droAna3.scaffold_13337 1845583 88 + 23293914 -------UUAAUCAUCC--UGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUG -------.........(--(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))))... ( -31.90, z-score = -2.10, R) >droEre2.scaffold_4784 13437001 84 - 25762168 -------------AUCACCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUG -------------..((.((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))).)) ( -32.30, z-score = -2.22, R) >droYak2.chr3L 13528204 85 - 24197627 ------------CUUCACCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUG ------------...((.((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))).)) ( -32.30, z-score = -2.20, R) >droSec1.super_0 5600852 87 - 21120651 ----------UUUAUCACCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUG ----------.....((.((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))).)) ( -32.30, z-score = -2.35, R) >droSim1.chr3L 12827177 87 - 22553184 ----------UUUAUCACCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUG ----------.....((.((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))).)) ( -32.30, z-score = -2.35, R) >anoGam1.chr2L 19552281 84 + 48795086 ----------AACAUCCAUUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACCG--- ----------..........((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).)))))))..--- ( -29.70, z-score = -1.09, R) >consensus ___________UCAUCACCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUG ...................(((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).)))))))).... (-29.48 = -28.58 + -0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:30 2011