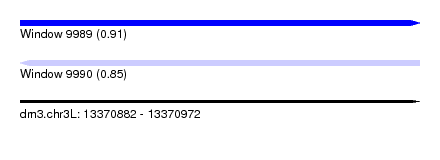
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,370,882 – 13,370,972 |
| Length | 90 |
| Max. P | 0.912227 |

| Location | 13,370,882 – 13,370,972 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 66.00 |
| Shannon entropy | 0.69037 |
| G+C content | 0.35198 |
| Mean single sequence MFE | -14.95 |
| Consensus MFE | -4.84 |
| Energy contribution | -4.86 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
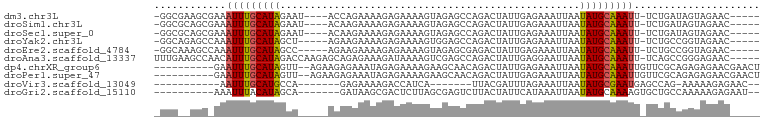
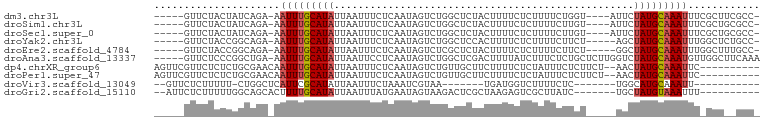
>dm3.chr3L 13370882 90 + 24543557 -GGCGAAGCGAAAUUUGCAUAGAAU----ACCAGAAAAGAGAAAAGUAGAGCCAGACUAUUGAGAAAUUAAUAUGCAAAUU-UCUGAUAGUAGAAC----- -.((.....((((((((((((....----...............(((........))).((((....))))))))))))))-)).....)).....----- ( -15.70, z-score = -2.88, R) >droSim1.chr3L 12768367 90 + 22553184 -GGCGCAGCGAAAUUUGCAUAGAAU----ACAAGAAAAGAGAAAAGUAGAGCCAGACUAUUGAGAAAUUAAUAUGCAAAUU-UCUGAUAGUAGAAC----- -(((((((......))))......(----((..............)))..)))..((((((.(((((((........))))-))))))))).....----- ( -16.34, z-score = -2.59, R) >droSec1.super_0 5545567 90 + 21120651 -GGCGCAGCGAAAUUUGCAUAGAAU----ACAAGAAAAGAGAAAAGUAGAGCCAGACUAUUGAGAAAUUAAUAUGCAAAUU-UCUGAUAGUAGAAC----- -(((((((......))))......(----((..............)))..)))..((((((.(((((((........))))-))))))))).....----- ( -16.34, z-score = -2.59, R) >droYak2.chr3L 13468216 89 + 24197627 -GGCAGAGCCAAAUUUGCAUAGCU-----AGAAGAAAAGAGAAAAGUGGAGCCAGACUAUUGAGAAAUUAAUAUGCAAAUU-UCUGCCGGUAGAAC----- -((((((....((((((((((...-----.................((....)).....((((....))))))))))))))-))))))........----- ( -21.20, z-score = -2.65, R) >droEre2.scaffold_4784 13383114 89 + 25762168 -GGCAAAGCCAAAUUUGCAUAGCC-----AGAAGAAAAGAGAAAAGUAGAGCGAGACUAUUGAGAAAUUAAUAUGCAAAUU-UCUGCCGGUAGAAC----- -((((.....(((((((((((...-----...............(((........))).((((....))))))))))))))-).))))........----- ( -16.70, z-score = -1.73, R) >droAna3.scaffold_13337 11177526 95 - 23293914 UUUGAAGCCAACAUUUGCAUAGACCAAGAGCAGAGAAAGAUAAAAGUCGAGCCAGACUAUUGAGGAAUUAAUAUGCAAAUU-UCAGCCGGGAGAAC----- ......((....(((((((((.......................((((......)))).((((....))))))))))))).-...)).........----- ( -13.40, z-score = -0.21, R) >dp4.chrXR_group6 5638263 89 + 13314419 ----------GAAUUUGCAUAGUU--AGAAGAGAAAUAGAGAAAAGAAGCAACAGACUAUUGAGAAAUUAAUAUGCAAAUUGUUCGCAGAGAGAACGAACU ----------.((((((((((.((--((......(((((.................)))))......))))))))))))))(((((.........))))). ( -14.73, z-score = -2.70, R) >droPer1.super_47 128845 89 + 592741 ----------GAAUUUGCAUAGUU--AGAAGAGAAAUAGAGAAAAGAAGCAACAGACUAUUGAGAAAUUAAUAUGCAAAUUGUUCGCAGAGAGAACGAACU ----------.((((((((((.((--((......(((((.................)))))......))))))))))))))(((((.........))))). ( -14.73, z-score = -2.70, R) >droVir3.scaffold_13049 12803980 73 + 25233164 -----------AAUUUGCAUGCCA-------GAGAAAAGACCAUCA-------UUACGAUUUAGAAAUUAAUAUGCGAAUGAGCCAG-AAAAAGAGAAC-- -----------.(((((((((...-------..((........)).-------....((((....))))..))))))))).......-...........-- ( -6.00, z-score = -0.57, R) >droGri2.scaffold_15110 23457834 82 + 24565398 ----------AAAUUUACAUAGCA-------GAUAAGCGACUCUUAGCGAGUCUUACUAUUCAUAAAUUAAUAUGCAAAAGUGCUGCCAAAAAGAGAAU-- ----------...........(((-------(......(((((.....))))).((((.((((((......)))).)).))))))))............-- ( -14.40, z-score = -1.04, R) >consensus _GGC__AGC_AAAUUUGCAUAGAA_____ACAAGAAAAGAGAAAAGUAGAGCCAGACUAUUGAGAAAUUAAUAUGCAAAUU_UCUGCCAGUAGAAC_____ ............(((((((((..................................................)))))))))..................... ( -4.84 = -4.86 + 0.02)
| Location | 13,370,882 – 13,370,972 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 66.00 |
| Shannon entropy | 0.69037 |
| G+C content | 0.35198 |
| Mean single sequence MFE | -14.04 |
| Consensus MFE | -3.90 |
| Energy contribution | -4.11 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
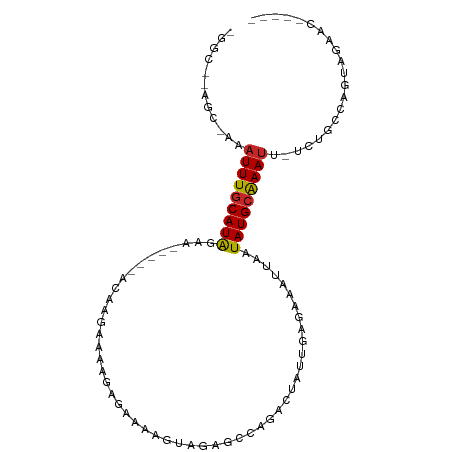
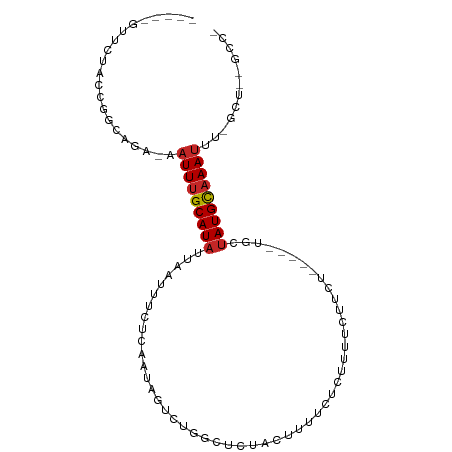
>dm3.chr3L 13370882 90 - 24543557 -----GUUCUACUAUCAGA-AAUUUGCAUAUUAAUUUCUCAAUAGUCUGGCUCUACUUUUCUCUUUUCUGGU----AUUCUAUGCAAAUUUCGCUUCGCC- -----............((-((((((((((........(((......)))...((((............)))----)...))))))))))))........- ( -13.30, z-score = -2.39, R) >droSim1.chr3L 12768367 90 - 22553184 -----GUUCUACUAUCAGA-AAUUUGCAUAUUAAUUUCUCAAUAGUCUGGCUCUACUUUUCUCUUUUCUUGU----AUUCUAUGCAAAUUUCGCUGCGCC- -----............((-((((((((((........(((......)))...(((..............))----)...))))))))))))........- ( -12.24, z-score = -1.60, R) >droSec1.super_0 5545567 90 - 21120651 -----GUUCUACUAUCAGA-AAUUUGCAUAUUAAUUUCUCAAUAGUCUGGCUCUACUUUUCUCUUUUCUUGU----AUUCUAUGCAAAUUUCGCUGCGCC- -----............((-((((((((((........(((......)))...(((..............))----)...))))))))))))........- ( -12.24, z-score = -1.60, R) >droYak2.chr3L 13468216 89 - 24197627 -----GUUCUACCGGCAGA-AAUUUGCAUAUUAAUUUCUCAAUAGUCUGGCUCCACUUUUCUCUUUUCUUCU-----AGCUAUGCAAAUUUGGCUCUGCC- -----........((((((-((((((((((.........((......))(((....................-----)))))))))))))....))))))- ( -16.45, z-score = -1.55, R) >droEre2.scaffold_4784 13383114 89 - 25762168 -----GUUCUACCGGCAGA-AAUUUGCAUAUUAAUUUCUCAAUAGUCUCGCUCUACUUUUCUCUUUUCUUCU-----GGCUAUGCAAAUUUGGCUUUGCC- -----........((((((-((((((((..............(((((.........................-----)))))))))))))....))))))- ( -13.44, z-score = -0.79, R) >droAna3.scaffold_13337 11177526 95 + 23293914 -----GUUCUCCCGGCUGA-AAUUUGCAUAUUAAUUCCUCAAUAGUCUGGCUCGACUUUUAUCUUUCUCUGCUCUUGGUCUAUGCAAAUGUUGGCUUCAAA -----........(((..(-.(((((((((......((..........))...((((...................))))))))))))).)..)))..... ( -16.81, z-score = -1.21, R) >dp4.chrXR_group6 5638263 89 - 13314419 AGUUCGUUCUCUCUGCGAACAAUUUGCAUAUUAAUUUCUCAAUAGUCUGUUGCUUCUUUUCUCUAUUUCUCUUCU--AACUAUGCAAAUUC---------- .((((((.......))))))(((((((((((((.......(((((.................))))).......)--)).)))))))))).---------- ( -15.97, z-score = -5.19, R) >droPer1.super_47 128845 89 - 592741 AGUUCGUUCUCUCUGCGAACAAUUUGCAUAUUAAUUUCUCAAUAGUCUGUUGCUUCUUUUCUCUAUUUCUCUUCU--AACUAUGCAAAUUC---------- .((((((.......))))))(((((((((((((.......(((((.................))))).......)--)).)))))))))).---------- ( -15.97, z-score = -5.19, R) >droVir3.scaffold_13049 12803980 73 - 25233164 --GUUCUCUUUUU-CUGGCUCAUUCGCAUAUUAAUUUCUAAAUCGUAA-------UGAUGGUCUUUUCUC-------UGGCAUGCAAAUU----------- --...........-...........((((............(((....-------.))).(((.......-------.))))))).....----------- ( -6.10, z-score = 0.51, R) >droGri2.scaffold_15110 23457834 82 - 24565398 --AUUCUCUUUUUGGCAGCACUUUUGCAUAUUAAUUUAUGAAUAGUAAGACUCGCUAAGAGUCGCUUAUC-------UGCUAUGUAAAUUU---------- --..........((((((.(((....((((......))))...)))..(((((.....)))))......)-------))))).........---------- ( -17.90, z-score = -1.66, R) >consensus _____GUUCUACCGGCAGA_AAUUUGCAUAUUAAUUUCUCAAUAGUCUGGCUCUACUUUUCUCUUUUCUUCU_____UGCUAUGCAAAUUU_GCU__GCC_ .....................(((((((((..................................................)))))))))............ ( -3.90 = -4.11 + 0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:27 2011