
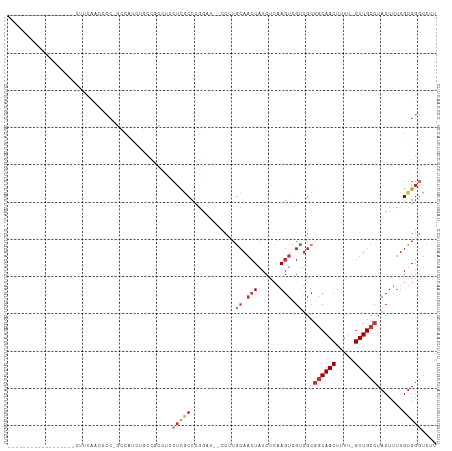
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 13,325,775 – 13,325,868 |
| Length | 93 |
| Max. P | 0.841620 |

| Location | 13,325,775 – 13,325,868 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 70.58 |
| Shannon entropy | 0.53586 |
| G+C content | 0.53303 |
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -9.45 |
| Energy contribution | -10.00 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.841620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13325775 93 + 24543557 ------------------CUUCAACGCC-GCCAUCUGCCGCCUUCUCGCCAGGAU--CCUUGCAACUAUCUCAAGUCGUUGCGGCAACUUUU-GUUGCCUACUUUUGUGGGUCUU ------------------.......((.-((.....)).)).........(((((--((..(((((.((.....)).)))))((((((....-)))))).........))))))) ( -25.40, z-score = -1.99, R) >droAna3.scaffold_13337 11135277 92 - 23293914 ------------------CGCUGUCCCC-GAUCCCCUUUUCUCUGUCCCCUGUCU--CCAUGCAACUAUCUCAAGUCGUUG-GGCAACUUUU-GUUGCCUACUUUUGUGGAUCCG ------------------..........-(((((.....................--....((.(((......))).))((-((((((....-)))))))).......))))).. ( -18.70, z-score = -2.33, R) >droEre2.scaffold_4784 13337757 93 + 25762168 ------------------CUACAAUCCC-GCCGUUUUCCGCCUCCUCGCCUGGAU--GCUUGCAACUAUCUCAAGUCGUUGCGGCAACUUUU-GUUGCCUACUUUUGUGGGUCUU ------------------.......(((-((........((.(((......))).--))..(((((.((.....)).)))))((((((....-)))))).......))))).... ( -28.10, z-score = -3.58, R) >droYak2.chr3L 13421974 93 + 24197627 ------------------CUCCAAUCCC-GCCGUUUGCCGCCUCCUCGCCUGGAU--CCUUGCAACUAUCUCAAGUCGCUGCGACAACUUUU-GUUGCCUACUUUUGUGGGUCUU ------------------.......(((-((...((((....(((......))).--....)))).......((((....((((((.....)-)))))..))))..))))).... ( -19.20, z-score = -0.80, R) >droSec1.super_0 5500406 93 + 21120651 ------------------CUUCAACGCC-GCCAUCUGCCGCCUUCUCGCCUGGAU--CCUUGCAACUAUCUCAAGUCGUUGCGGCAACUUUU-GUUGCCUACUUUUGUGGGUCUU ------------------.......((.-((.....)).))..........((((--((..(((((.((.....)).)))))((((((....-)))))).........)))))). ( -24.80, z-score = -2.06, R) >droSim1.chrU 6680110 93 - 15797150 ------------------CUUCAACGCC-GCCAUCUGCCGCCUUCUCGCCUGGAU--CCUUGCAACUAUCUCAAGUCGUUGCGGCAACUUUU-GUUGCCUACUUUUGUGGGUCUU ------------------.......((.-((.....)).))..........((((--((..(((((.((.....)).)))))((((((....-)))))).........)))))). ( -24.80, z-score = -2.06, R) >dp4.chrXR_group6 7728317 114 - 13314419 CUGCACCUCUGCACCGCCCAUUACCCCCCAAUGCCUUGCCUGUCCUUGCCAAUCUGCCCCUGCAACUAUCUCAAGUCGCUGCGGCAACUUUU-GUUGCCUACUUUUGUGGGUCCU .((((....))))..((((((................((..((.((((......(((....))).......))))..)).))((((((....-)))))).......))))))... ( -25.22, z-score = -1.50, R) >droVir3.scaffold_13049 12486196 75 - 25233164 ------------------------------AGCCACGCCCUGCUCAUACCUUGCCGCCCCUA----UAUCUGCAGUUGUCGCCGCAACUUUUCGUUGCCUACUUAUGUG------ ------------------------------.((.(((..((((...................----.....)))).))).)).(((((.....)))))...........------ ( -11.56, z-score = -0.38, R) >consensus __________________CUUCAACCCC_GCCAUCUGCCGCCUUCUCGCCUGGAU__CCUUGCAACUAUCUCAAGUCGUUGCGGCAACUUUU_GUUGCCUACUUUUGUGGGUCUU ............................................(((((............((.(((......))).))...((((((.....)))))).......))))).... ( -9.45 = -10.00 + 0.55)


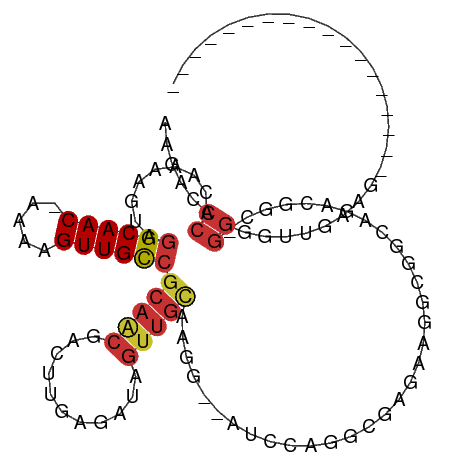
| Location | 13,325,775 – 13,325,868 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 70.58 |
| Shannon entropy | 0.53586 |
| G+C content | 0.53303 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -11.41 |
| Energy contribution | -11.93 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 13325775 93 - 24543557 AAGACCCACAAAAGUAGGCAAC-AAAAGUUGCCGCAACGACUUGAGAUAGUUGCAAGG--AUCCUGGCGAGAAGGCGGCAGAUGGC-GGCGUUGAAG------------------ ..(((((.(.......((((((-....))))))(((((...........)))))....--...(((.((......)).)))...).-)).)))....------------------ ( -25.80, z-score = -0.82, R) >droAna3.scaffold_13337 11135277 92 + 23293914 CGGAUCCACAAAAGUAGGCAAC-AAAAGUUGCC-CAACGACUUGAGAUAGUUGCAUGG--AGACAGGGGACAGAGAAAAGGGGAUC-GGGGACAGCG------------------ ..(((((.(..((((.((((((-....))))))-.....))))..)...(((.(.((.--...)).).)))..........)))))-(....)....------------------ ( -27.00, z-score = -3.58, R) >droEre2.scaffold_4784 13337757 93 - 25762168 AAGACCCACAAAAGUAGGCAAC-AAAAGUUGCCGCAACGACUUGAGAUAGUUGCAAGC--AUCCAGGCGAGGAGGCGGAAAACGGC-GGGAUUGUAG------------------ ....(((.(.......((((((-....))))))(((((...........)))))..((--.(((......))).))........).-))).......------------------ ( -27.20, z-score = -2.12, R) >droYak2.chr3L 13421974 93 - 24197627 AAGACCCACAAAAGUAGGCAAC-AAAAGUUGUCGCAGCGACUUGAGAUAGUUGCAAGG--AUCCAGGCGAGGAGGCGGCAAACGGC-GGGAUUGGAG------------------ ....(((.(.......(....)-.....(((((((.((((((......))))))....--.(((......))).)))))))...).-))).......------------------ ( -27.40, z-score = -1.93, R) >droSec1.super_0 5500406 93 - 21120651 AAGACCCACAAAAGUAGGCAAC-AAAAGUUGCCGCAACGACUUGAGAUAGUUGCAAGG--AUCCAGGCGAGAAGGCGGCAGAUGGC-GGCGUUGAAG------------------ ..((.((.........((((((-....))))))(((((...........)))))..))--.))........((.((.((.....))-.)).))....------------------ ( -25.30, z-score = -1.05, R) >droSim1.chrU 6680110 93 + 15797150 AAGACCCACAAAAGUAGGCAAC-AAAAGUUGCCGCAACGACUUGAGAUAGUUGCAAGG--AUCCAGGCGAGAAGGCGGCAGAUGGC-GGCGUUGAAG------------------ ..((.((.........((((((-....))))))(((((...........)))))..))--.))........((.((.((.....))-.)).))....------------------ ( -25.30, z-score = -1.05, R) >dp4.chrXR_group6 7728317 114 + 13314419 AGGACCCACAAAAGUAGGCAAC-AAAAGUUGCCGCAGCGACUUGAGAUAGUUGCAGGGGCAGAUUGGCAAGGACAGGCAAGGCAUUGGGGGGUAAUGGGCGGUGCAGAGGUGCAG ....(((......((.((((((-....)))))))).((((((......)))))).)))(((..((.((........))...((((((............)))))).))..))).. ( -32.90, z-score = -1.03, R) >droVir3.scaffold_13049 12486196 75 + 25233164 ------CACAUAAGUAGGCAACGAAAAGUUGCGGCGACAACUGCAGAUA----UAGGGGCGGCAAGGUAUGAGCAGGGCGUGGCU------------------------------ ------...........(((((.....)))))(((.((..((((..(((----(...(....)...))))..))))...)).)))------------------------------ ( -19.50, z-score = -1.45, R) >consensus AAGACCCACAAAAGUAGGCAAC_AAAAGUUGCCGCAACGACUUGAGAUAGUUGCAAGG__AUCCAGGCGAGAAGGCGGCAGACGGC_GGGGUUGAAG__________________ .....((.........((((((.....))))))(((((...........)))))...........))................................................ (-11.41 = -11.93 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:23:21 2011